26.11: Enzymes and Coenzymes
- Page ID
- 36476
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)After completing this section, you should be able to
- describe the catalytic role of an enzyme in a biochemical reaction.
- give an example of one fat‑soluble and one water‑soluble vitamin.
Make certain that you can define, and use in context, the key term below.
- coenzyme
- cofactor
- enzyme
- substrate
- vitamin
You should have a general knowledge of the function of enzymes, but you need not memorize specific names or the classification system.
A catalyst is any substance that increases the rate or speed of a chemical reaction without being changed or consumed in the reaction. Enzymes are biological catalysts, and nearly all of them are proteins. In addition, enzymes are highly specific in their action; that is, each enzyme catalyzes only one type of reaction in only one compound or a group of structurally related compounds. The compound or compounds on which an enzyme acts are known as its substrates. Enzymes are classified by reaction type into six categories show in Table \(\PageIndex{1}\).
| Class | Type of Reaction Catalyzed | Examples |
|---|---|---|
| oxidoreductases | oxidation-reduction reactions | Dehydrogenases catalyze oxidation-reduction reactions involving hydrogen and reductases catalyze reactions in which a substrate is reduced. |
| transferases | transfer reactions of groups, such as methyl, amino, and acetyl | Transaminases catalyze the transfer of amino group, and kinases catalyze the transfer of a phosphate group. |
| hydrolases | hydrolysis reactions | Lipases catalyze the hydrolysis of lipids, and proteases catalyze the hydrolysis of proteins |
| lyases | reactions in which groups are removed without hydrolysis or addition of groups to a double bond | Decarboxylases catalyze the removal of carboxyl groups. |
| isomerases | reactions in which a compound is converted to its isomer | Isomerases may catalyze the conversion of an aldose to a ketose, and mutases catalyze reactions in which a functional group is transferred from one atom in a substrate to another. |
| ligases | reactions in which new bonds are formed between carbon and another atom; energy is required | Synthetases catalyze reactions in which two smaller molecules are linked to form a larger one. |
Enzyme-catalyzed reactions occur in at least two steps. In the first step, an enzyme molecule (E) and the substrate molecule or molecules (S) collide and react to form an intermediate compound called the enzyme-substrate (E–S) complex (Equation \(\ref{step1}\)). This step is reversible because the complex can break apart into the original substrate or substrates and the free enzyme. Once the E–S complex forms, the enzyme is able to catalyze the formation of product (P), which is then released from the enzyme surface (Equation \(\ref{step2}\)):
\[S + E \rightleftharpoons E–S \label{step1} \]
\[E–S → P + E \label{step2} \]
Hydrogen bonding and other electrostatic interactions hold the enzyme and substrate together in the complex. The structural features or functional groups on the enzyme that participate in these interactions are located in a cleft or pocket on the enzyme surface. This pocket, where the enzyme combines with the substrate and transforms the substrate to product is called the active site of the enzyme (Figure \(\PageIndex{1}\)).
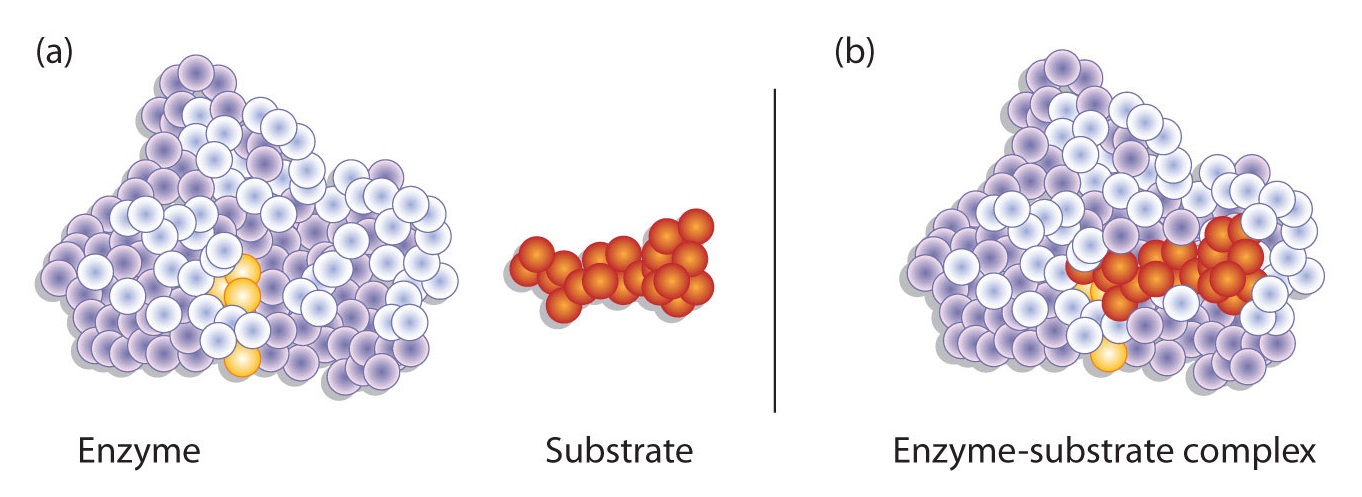
Figure \(\PageIndex{1}\): Substrate Binding to the Active Site of an Enzyme. The enzyme dihydrofolate reductase is shown with one of its substrates: NADP+ (a) unbound and (b) bound. The NADP+ (shown in red) binds to a pocket that is complementary to it in shape and ionic properties.
The active site possesses a unique conformation (including correctly positioned bonding groups) that is complementary to the structure of the substrate, so that the enzyme and substrate molecules fit together in much the same manner as a key fits into a tumbler lock. In fact, an early model describing the formation of the enzyme-substrate complex was called the lock-and-key model (Figure \(\PageIndex{2}\)). This model portrayed the enzyme as conformationally rigid and able to bond only to substrates that exactly fit the active site.
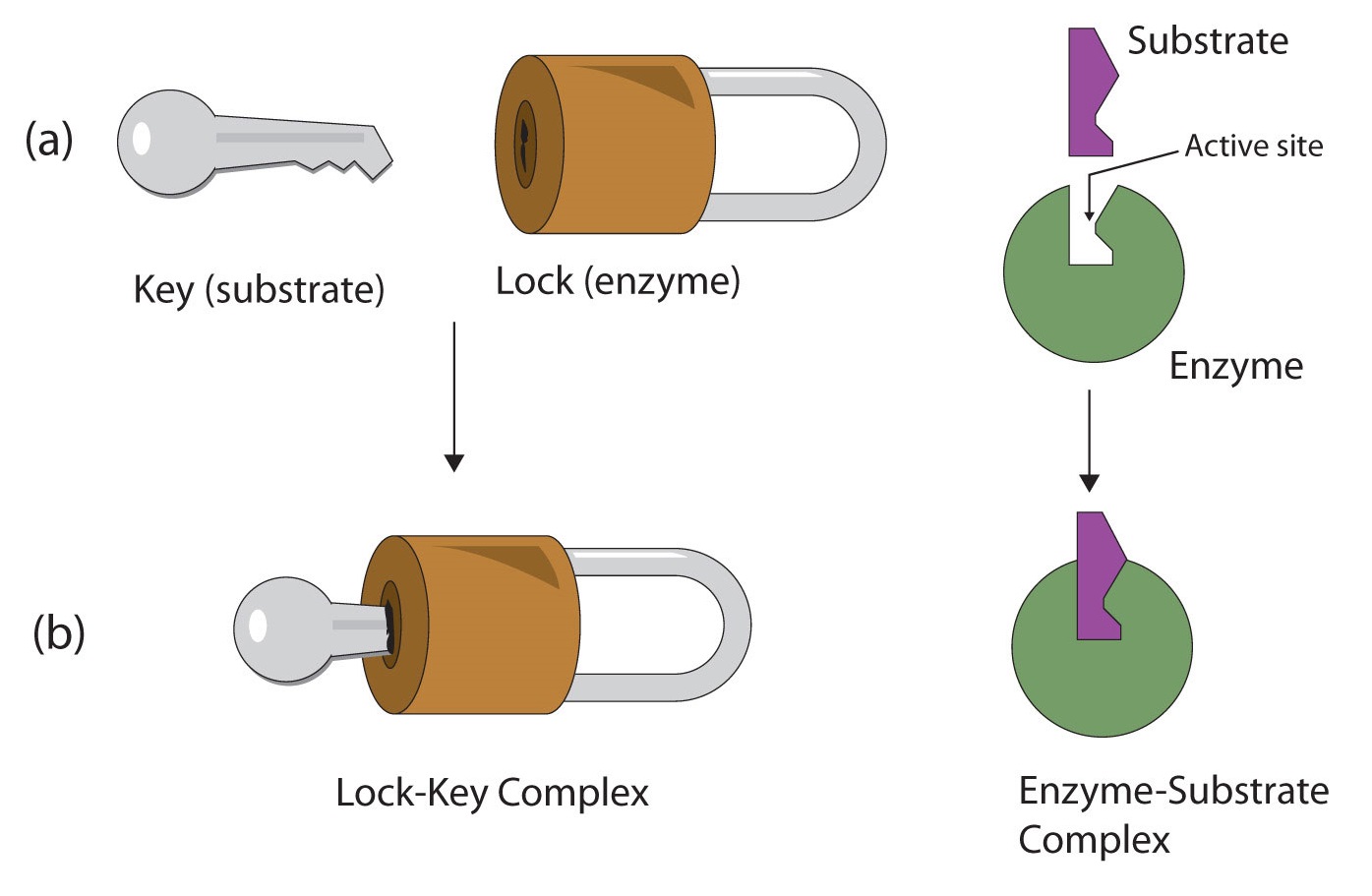
Working out the precise three-dimensional structures of numerous enzymes has enabled chemists to refine the original lock-and-key model of enzyme actions. They discovered that the binding of a substrate often leads to a large conformational change in the enzyme, as well as to changes in the structure of the substrate or substrates. The current theory, known as theinduced-fit model, says that enzymes can undergo a change in conformation when they bind substrate molecules, and the active site has a shape complementary to that of the substrate only after the substrate is bound, as shown for hexokinase in Figure \(\PageIndex{3}\). After catalysis, the enzyme resumes its original structure.
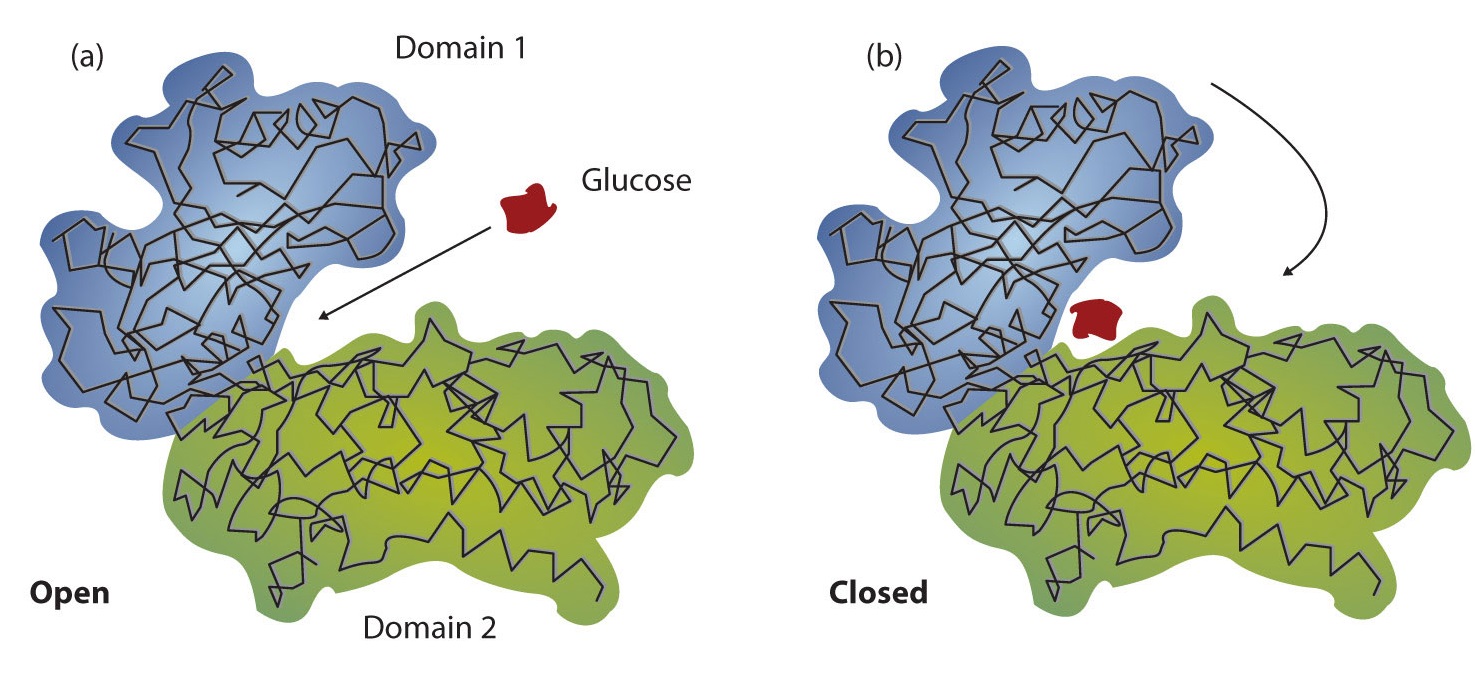
The structural changes that occur when an enzyme and a substrate join together bring specific parts of a substrate into alignment with specific parts of the enzyme’s active site. Amino acid side chains in or near the binding site can then act as acid or base catalysts, provide binding sites for the transfer of functional groups from one substrate to another or aid in the rearrangement of a substrate. The participating amino acids, which are usually widely separated in the primary sequence of the protein, are brought close together in the active site as a result of the folding and bending of the polypeptide chain or chains when the protein acquires its tertiary and quaternary structure. Binding to enzymes brings reactants close to each other and aligns them properly, which has the same effect as increasing the concentration of the reacting compounds.
- What type of interaction would occur between an OH group present on a substrate molecule and a functional group in the active site of an enzyme?
- Suggest an amino acid whose side chain might be in the active site of an enzyme and form the type of interaction you just identified.
Solution
- An OH group would most likely engage in hydrogen bonding with an appropriate functional group present in the active site of an enzyme.
- Several amino acid side chains would be able to engage in hydrogen bonding with an OH group. One example would be asparagine, which has an amide functional group.
- What type of interaction would occur between an COO− group present on a substrate molecule and a functional group in the active site of an enzyme?
- Suggest an amino acid whose side chain might be in the active site of an enzyme and form the type of interaction you just identified.
Enzyme Cofactors and Vitamins
Many enzymes are simple proteins consisting entirely of one or more amino acid chains. Other enzymes contain a nonprotein component called a cofactor that is necessary for the enzyme’s proper functioning. There are two types of cofactors: inorganic ions [e.g., zinc or Cu(I) ions] and organic molecules known as coenzymes. Most coenzymes are vitamins or are derived from vitamins.
Vitamins are organic compounds that are essential in very small (trace) amounts for the maintenance of normal metabolism. They generally cannot be synthesized at adequate levels by the body and must be obtained from the diet. The absence or shortage of a vitamin may result in a vitamin-deficiency disease. In the first half of the 20th century, a major focus of biochemistry was the identification, isolation, and characterization of vitamins. Despite accumulating evidence that people needed more than just carbohydrates, fats, and proteins in their diets for normal growth and health, it was not until the early 1900s that research established the need for trace nutrients in the diet.
| Vitamin | Physiological Function | Effect of Deficiency |
|---|---|---|
| vitamin A (retinol) | formation of vision pigments; differentiation of epithelial cells | night blindness; continued deficiency leads to total blindness |
| vitamin D (cholecalciferol) | increases the body’s ability to absorb calcium and phosphorus | osteomalacia (softening of the bones); known as rickets in children |
| vitamin E (tocopherol) | fat-soluble antioxidant | damage to cell membranes |
| vitamin K (phylloquinone) | formation of prothrombin, a key enzyme in the blood-clotting process | increases the time required for blood to clot |
Because organisms differ in their synthetic abilities, a substance that is a vitamin for one species may not be so for another. Over the past 100 years, scientists have identified and isolated 13 vitamins required in the human diet and have divided them into two broad categories: the fat-soluble vitamins (Table \(\PageIndex{2}\)), which include vitamins A, D, E, and K, and the water-soluble vitamins, which are the B complex vitamins and vitamin C (Table \(\PageIndex{3}\)). All fat-soluble vitamins contain a high proportion of hydrocarbon structural components. There are one or two oxygen atoms present, but the compounds as a whole are nonpolar. In contrast, water-soluble vitamins contain large numbers of electronegative oxygen and nitrogen atoms, which can engage in hydrogen bonding with water. Most water-soluble vitamins act as coenzymes or are required for the synthesis of coenzymes. The fat-soluble vitamins are important for a variety of physiological functions.
| Vitamin | Coenzyme | Coenzyme Function | Deficiency Disease |
|---|---|---|---|
| vitamin B1 (thiamine) | thiamine pyrophosphate | decarboxylation reactions | beri-beri |
| vitamin B2 (riboflavin) | flavin mononucleotide or flavin adenine dinucleotide | oxidation-reduction reactions involving two hydrogen atoms | — |
| vitamin B3 (niacin) | nicotinamide adenine dinucleotide or nicotinamide adenine dinucleotide phosphate | oxidation-reduction reactions involving the hydride ion (H−) | pellagra |
| vitamin B6 (pyridoxine) | pyridoxal phosphate | variety of reactions including the transfer of amino groups | — |
| vitamin B12 (cyanocobalamin) | methylcobalamin or deoxyadenoxylcobalamin | intramolecular rearrangement reactions | pernicious anemia |
| biotin | biotin | carboxylation reactions | — |
| folic acid | tetrahydrofolate | carrier of one-carbon units such as the formyl group | anemia |
| pantothenic Acid | coenzyme A | carrier of acyl groups | — |
| vitamin C (ascorbic acid) | none | antioxidant; formation of collagen, a protein found in tendons, ligaments, and bone | scurvy |
One characteristic that distinguishes an enzyme from all other types of catalysts is its substrate specificity. An inorganic acid such as sulfuric acid can be used to increase the reaction rates of many different reactions, such as the hydrolysis of disaccharides, polysaccharides, lipids, and proteins, with complete impartiality. In contrast, enzymes are much more specific. Some enzymes act on a single substrate, while other enzymes act on any of a group of related molecules containing a similar functional group or chemical bond. Some enzymes even distinguish between D- and L-stereoisomers, binding one stereoisomer but not the other. Urease, for example, is an enzyme that catalyzes the hydrolysis of a single substrate—urea—but not the closely related compounds methyl urea, thiourea, or biuret. The enzyme carboxypeptidase, on the other hand, is far less specific. It catalyzes the removal of nearly any amino acid from the carboxyl end of any peptide or protein.
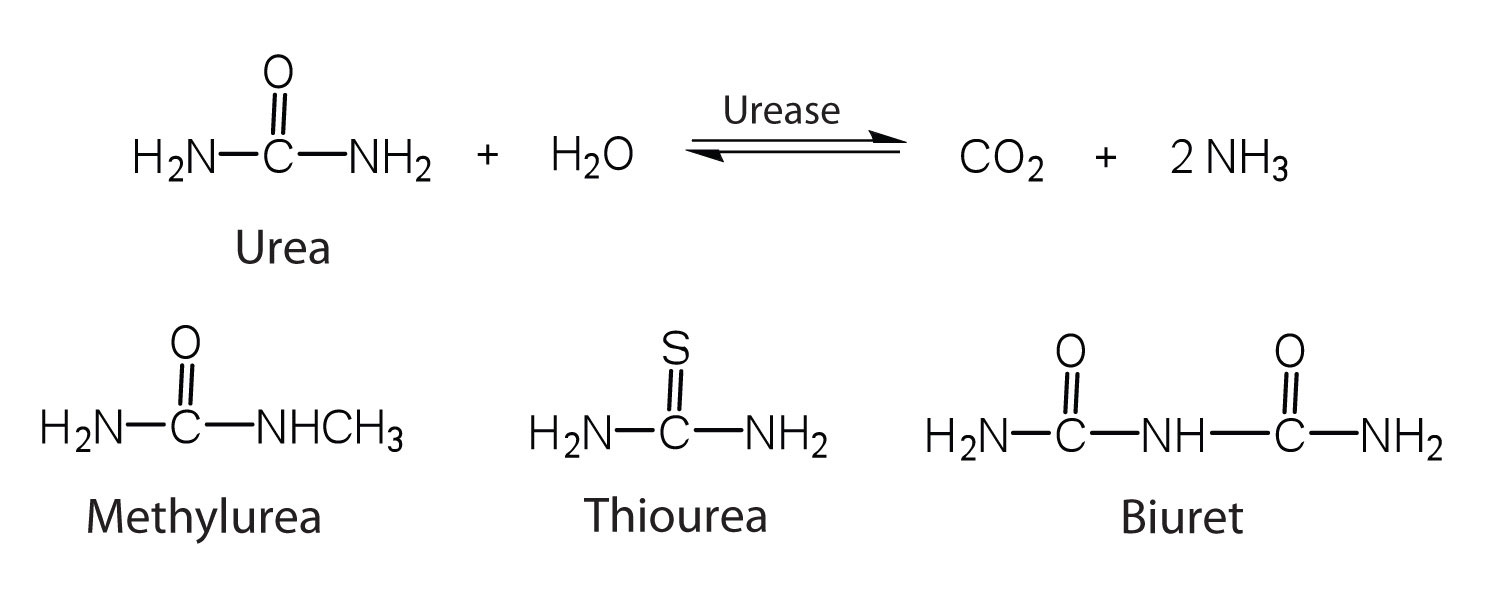
Enzyme specificity results from the uniqueness of the active site in each different enzyme because of the identity, charge, and spatial orientation of the functional groups located there. It regulates cell chemistry so that the proper reactions occur in the proper place at the proper time. Clearly, it is crucial to the proper functioning of the living cell.
Concept Review Exercises
-
Distinguish between the lock-and-key model and induced-fit model of enzyme action.
-
Which enzyme has greater specificity—urease or carboxypeptidase? Explain.
Answers
-
The lock-and-key model portrays an enzyme as conformationally rigid and able to bond only to substrates that exactly fit the active site. The induced fit model portrays the enzyme structure as more flexible and is complementary to the substrate only after the substrate is bound.
-
Urease has the greater specificity because it can bind only to a single substrate. Carboxypeptidase, on the other hand, can catalyze the removal of nearly any amino acid from the carboxyl end of a peptide or protein.
Takeaways
- A substrate binds to a specific region on an enzyme known as the active site, where the substrate can be converted to product.
- The substrate binds to the enzyme primarily through hydrogen bonding and other electrostatic interactions.
- The induced-fit model says that an enzyme can undergo a conformational change when binding a substrate.
- Enzymes exhibit varying degrees of substrate specificity.
Exercises
-
What type of interaction would occur between each group present on a substrate molecule and a functional group of the active site in an enzyme?
- COOH
- NH3+
- OH
- CH(CH3)2
-
What type of interaction would occur between each group present on a substrate molecule and a functional group of the active site in an enzyme?
- SH
- NH2
- C6H5
- COO−
-
For each functional group in Exercise 1, suggest an amino acid whose side chain might be in the active site of an enzyme and form the type of interaction you identified.
-
For each functional group in Exercise 2, suggest an amino acid whose side chain might be in the active site of an enzyme and form the type of interaction you identified.
Answers
-
- hydrogen bonding
- ionic bonding
- hydrogen bonding
- dispersion forces
-
- The amino acid has a polar side chain capable of engaging in hydrogen bonding; serine (answers will vary).
- The amino acid has a negatively charged side chain; aspartic acid (answers will vary).
- The amino acid has a polar side chain capable of engaging in hydrogen bonding; asparagine (answers will vary).
- The amino acid has a nonpolar side chain; isoleucine (answers will vary).

