7.9: Enzymes
- Page ID
- 425778
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Understand enzyme-related terminology, nomenclature, and classification of enzymes.
- Understand the mode of action of enzymes, the factors that affect them, and the inhibitors that retard or damage the enzyme activity.
Enzymes and the related terminology
Three conditions are necessary for a chemical reaction to happen:
- Reactants must collide -the more frequent the collisions faster the reaction,
- the reactant must have proper orientation at the time of the collision -the higher the probability of proper orientation at the time of the collision, the faster the reaction, and
- there must be enough energy at collision to surpass the energy barrier, i.e., the energy of activation for the reaction -the lower the activation energy, the faster the reaction.
Often chemical reactions are possible but so slow that they are practically useless. For example, a reaction between hydrogen \(\ce{H2}\) and nitrogen \(\ce{N2}\) producing ammonia \(\ce{NH3}\) is possible, but to make it practically useful, high-pressure, high-temperature, and catalysts are needed.
\(\ce{N2(g) + H2(g) <=>[heat, pressure, catalyst] 2NH3(g)\;\; -92.4 kJ mol^{-1}}\)
A catalyst is a reagent that increases the rate of a chemical reaction without itself being altered in the process.
The catalysts usually increase the rate of chemical reactions by improving the last two factors, i.e., increasing the probability of proper orientation and providing an alternate route for the reaction with lower activation energy. There are more constraints for chemical reactions in living things, e.g., the reaction has to occur under physiological conditions of pH ~7.4 and body temperature ~37 oC. Special catalysts called enzymes are used to regulate chemical reactions in living things.
An enzyme is a substance that regulates the rate of chemical reaction in living things without itself being altered in the process. In other words, enzymes are biological catalysts.
Like other catalysts, the enzymes usually increase the rate of chemical reactions by improving the last two factors, i.e., increasing the probability of proper orientation and providing an alternate route for the reaction with lower activation energy. The enzymes achieve the appropriate orientation of the reactants by binding them in a specific region within the enzyme, which has the geometry of its interacting groups right for securing the reactant in a particular orientation.
A reactant in an organic or biochemical reaction is a substrate.
The enzyme's active site is the region within an enzyme where the substrate binds for the reaction.
Enzymes are usually proteins having primary, secondary, and tertiary structures. The active site is usually a small region (10% to 20%) within the enzyme. A few side chains of amino acid residues within the active site participate in the catalytic action, as shown in Figure \(\PageIndex{1}\). The rest of the amino acid residues define and hold the secondary and tertiary structures.

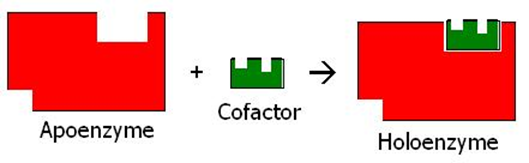
Some enzymes need a non-protein part to combine with them for their function.
The enzymes that need a non-protein portion to combine with them for their function are called apoenzymes. The non-protein portion of the enzymes is called the cofactor, as illustrated in Figure \(\PageIndex{2}\). The cofactor could be a metallic ion, .e.g., \(\ce{Zn^2+}\) or \(\ce{Mg^2+}\), or an organic compound. Organic cofactor is called coenzyme. The apoenzyme and cofactors together are called holoenzymes, as illustrated in Figure \(\PageIndex{2}\).
Enzymes usually make the reaction happen millions of times faster. For example, one molecule of carbonic anhydrase enzyme, shown in Figure \(\PageIndex{1}\), can catalyze about one million molecules in one second in human blood by the following reaction.

Copyright: Synpath, CC0, via Wikimedia Commons.
Overall reaction: \(\ce{CO2 + H2O <=> HCO3^{-} + H^{+}}\)
A catalyst or a reagent is selective if it produces one product preferentially or exclusively when more than one product is possibly formed.
Enzymes are not only selective; they usually react with one compound or stereoisomer.
Stereospecific catalysts or reagents are those that, when reacting with one stereoisomer, selectively produce a stereoisomer and either do not react with the isomers of the reactant or selectively produce other stereoisomers from them. Enzymes are usually stereospecific, i.e., they are selective among reactants and selective among products.
For example, the enzyme arginase is stereospecific, hydrolysis amino acid L-arginine to L-ornithine and urea, but does not react with D-arginine.

Usually, enzymes react with one compound or a specific bond of one compound. For example, enzyme ureas catalysis hydrolysis of urea (\(\ce{(NH2)2C=O}\)) and does not hydrolyze any other amide.
\[\ce{(NH2)2C=O + H2O ->[urease] 2NH3 + CO2}\nonumber \]
Trypsin is a digestive enzyme that cleaves peptide bonds of proteins but not every peptide bond, only those on the C-side of lysine and arginine residue.

Some enzymes are specific for a class of compounds, e.g., lipases catalyzing triglyceride hydrolysis, but do not react with carbohydrates or proteins.
Names and classification of enzymes
Names of enzymes are derived by replacing the end of the name of a reactant or reaction with the suffix ase. For example, sucrase hydrolysis sucrose, lipase hydrolyzes lipids, oxidase catalyzes oxidation reactions, dehydrogenase removes hydrogen atoms, etc. Some old names of enzymes have the suffix in, e.g., digestive enzymes pepsin and trypsin. In the recent classification of enzymes, the name or the class name indicates the type of reaction it catalyzes. There are six major classes of enzymes, as described in the following.
Oxidoreductases
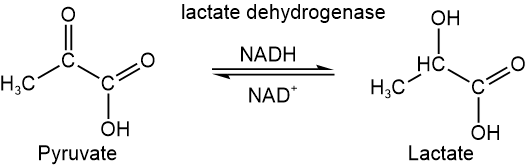
Oxidoreductases do oxidation-reduction reactions, i.e., oxidases oxidize a substance, reductases reduce a substance, and dehydrogenases remove hydrogen. For example, lactate dehydrogenase reduces pyruvate and oxidizes lactate, as shown below.

Transferases
Transferases transfer a group between two compounds. For example, kinases transfer phosphate groups, and transaminases transfer amino groups, as shown below.

Hydrolases
Hydrolases do hydrolysis reactions: lipases hydrolyze lipids, carbohydrates hydrolyze carbohydrates, proteases hydrolyze proteins, phosphatases hydrolyze phosphate esters, and nucleases hydrolyze nucleic acids. For example, acetylcholinesterase hydrolysis acetylcholine, as shown below.

Lyases
Lyases add two groups to double bond or remove two groups from adjacent atoms to create a double bond or a ring structure by means other than hydrolysis and oxidation, e.g., carboxylases add or remove CO2, and deaminases add or remove NH3. For example, actonitase, shown in Figure \(\PageIndex{3}\), adds or removes water. double bond or add a new ring structure.

Isomerases
Isomerases rearrange atoms in a molecule: isomerases convert cis to trans or trans to cis isomer, and epimerases convert D to L or L to D streoisomers. Phosphohexose isomerizes shown below isomerizes glucose-6-phosphate to fructose-6-phosphate

Ligases
Ligases or synthetases catalyze the joining of two molecules, e.g., glutamate to glutamine conversion by glutamine synthetase shown below.

(Copyright; Hbf878, CC0, via Wikimedia Commons)
How do enzymes catalyze the reactions?
The chemical reactions that enzymes catalyze can occur without enzymes, but the reactions' rates are usually prolonged due to high activation energies. Enzymes provide alternate routes to the reactions with lower energy barriers (activation energies) and proper orientations of substrates that result in fast reactions, as illustrated in Figure \(\PageIndex{3}\)

In a generalized enzyme-catalyzed reaction, enzyme (E) and substrate (S) bind to make an enzyme-substrate complex (ES). The intermolecular interactions between the enzyme and the substrate usually loosen the bonds of the substrate that need to be broken, resulting in a lower energy barrier for the catalyzed reaction that leads to the product (P) at a faster rate than otherwise. The product leaves the enzyme so it can bind with another substrate.
\[\ce{E + S <=> ES -> E + P}\nonumber \]
For example, carbonic anhydrase catalyzes the reaction of water (\(\ce{H2O}\)) with carbon dioxide (\(\ce{CO2}\)) in blood as illustrated in Figure \(\PageIndex{4}\). The carbonic anhydrase enzyme is bound with cofactor \(\ce{Zn^{2+}}\) through three histidine residues. The \(\ce{Zn^{2+}}\) is also bound with \(\ce{H2O}\) that bond makes water ready for the proton transfer. After the proton transfer, a strong electrophile \({-\overset{-}{O}H}\) is generated that attacks \(\ce{CO2}\), which is placed in the proper location in the hydrophobic pocket nearby. The product is displaced with another water molecule that repeats the process. One molecule of carbonic anhydrase can convert about million \(\ce{CO2}\) molecules per second in this reaction.


Models of enzyme action
Enzymes are usually specific for a particular substrate or a class of reaction. This specificity was explained first by models of enzyme action explained below.
Lock-and-key model
 The lock and key model assumes enzymes have a fixed shape with active sites similar to a lock in which a particular substrate with a fixed shape similar to a key can fit, as shown in the figure on the right.
The lock and key model assumes enzymes have a fixed shape with active sites similar to a lock in which a particular substrate with a fixed shape similar to a key can fit, as shown in the figure on the right.
This model explains the specificity of enzymes but ignores the dynamic nature of molecules. Further, according to this model, the products would be expected to be snug-fit in the active site and not easy to release. In reality, all the bonds in a molecule are vibrating and stretching, and the whole molecule is jiggling due to thermal energy, as illustrated in Figure \(\PageIndex{5}\).

Induced-fit model
The induced-fit model of enzyme action accounts for the flexibility of enzyme and substrate molecules. According to this model, the flexibility of the enzyme molecule allows the active site to adapt to the shape of the substrate and, at the same time, the substrate adopts the shape of the enzyme to acquire the best possible orientation for the reaction to occur, as illustrated in Figure \(\PageIndex{6}\). Then the active site shape re-adjusts to let the products be released to allow the next cycle of the enzyme action. The observation from experiments during the actual catalysis reaction supports the view that not only does the active site of the enzyme change shape, the backbone and the side chains of the enzyme molecule remain in constant motion during the enzyme action.

Experimental data shows that the active site is usually a tiny portion (10% to 20%) of the enzyme. Within the active site, two or a few side chains of amino acid residues usually catalyze the reaction. Usually, the catalytically active residue is one of the following: His, Cys, Asp, Arg, and Glu. These amino acids have acidic or basic or thiol functional groups, which are not only capable of hydrogen bonding but also capable of acid, base, electrophile, or nucleophile catalysis. For example, the pepsin enzyme breaks peptide bonds by using histidine and cystine, as illustrated in Figure \(\PageIndex{7}\).

The following video from RCSBProteinDataBank explains how enzymes work.
Isoenzymes are different enzymes that perform the same function in different body parts. They usually have a tertiary structure and differ in only a few amino acid residues. For example, lactate dehydrogenase (LDA) catalyzes the conversion between pyruvate and lactate, as shown below.
LDA is a tetramer made of two sub-units, the H-form, and the M-form, in five combinations found in different tissues, as shown in Table 1. Similarly, creatine phosphokinase (CPK) catalyzes the interconversion of phosphocreatine to creatine. CPK is a dimer in 3 isoenzyme forms shown in Table 2.
These isoenzymes usually function within cells. However, when some disease damages a tissue, the cells die, and the isoenzymes are released into the blood. Analysis of blood serum is used to diagnose the location of the damage. For example, an elevated level of LDH5 indicates liver damage and myocardial infarction (heart damage) is characterized by a high level of LDH1 isoenzyme. The heart damage will also elevate CK2.
| Type | Subunits | Illustration | Location |
|---|---|---|---|
| LDH1 | HHHH |  |
Heart and Erythrocyte |
| LDH2 | HHHM |  |
Heart and Erythrocyte |
| LDH3 | HHMM |  |
Brain and Kidney |
| LDH4 | HMMM |  |
Skeletal Muscle and Liver |
| LDH5 | MMMM |  |
Skeletal Muscle and Liver |
| Isoenzyme | Subunit | Illustration | Tissue of Origin |
|---|---|---|---|
| CPK1 | BB |  |
Brain |
| CPK2 | MB |  |
Heart |
| CPK3 | MM |  |
Skeletal muscle |
Factors that affect enzyme activity
The enzyme activity is related to how much it increases the reaction rate compared to the same reaction without the enzyme. The factors that affect enzyme activity include concentration, temperature, pH, and the presence of inhibitors.
Concentration
Usually, the enzyme is in very low concentration relative to the concentration of the substrate. Therefore,  an increase in the enzyme concentration increases the reaction rate
an increase in the enzyme concentration increases the reaction rate  linearly, i.e., doubling the enzyme concentration doubles the reaction rate, and tripling it triples the rate. An increase in the substrate concentration increases the reaction rate but not linearly; it follows a curvilinear curve. The rate increase reaches a saturation level and does not increase after that with a further increase in the substrate concentration, as illustrated in the figure on the left. This is because all the enzyme's active sites become occupied with the substrate at the saturation point, and the reaction proceeds at its maximum rate, as illustrated in the figure on the right.
linearly, i.e., doubling the enzyme concentration doubles the reaction rate, and tripling it triples the rate. An increase in the substrate concentration increases the reaction rate but not linearly; it follows a curvilinear curve. The rate increase reaches a saturation level and does not increase after that with a further increase in the substrate concentration, as illustrated in the figure on the left. This is because all the enzyme's active sites become occupied with the substrate at the saturation point, and the reaction proceeds at its maximum rate, as illustrated in the figure on the right.
Temperature
 Generally, an increase in temperature increases the rate of a chemical reaction. This is because more molecules have energy than activation energy at higher temperatures. The same applies to enzyme-catalyzed reactions at lower temperature ranges. Still, the rate becomes optimum at around body temperature and then starts to fall off, as illustrated in the figure on the right. The enzymes have secondary, tertiary, and quaternary structures optimized to perform best at the body temperature of about 37 oC. Denaturation inactivates the enzymes. It is reversible if the temperature is slightly above body temperature, but enzymes are denatured beyond repair at a much higher temperature.
Generally, an increase in temperature increases the rate of a chemical reaction. This is because more molecules have energy than activation energy at higher temperatures. The same applies to enzyme-catalyzed reactions at lower temperature ranges. Still, the rate becomes optimum at around body temperature and then starts to fall off, as illustrated in the figure on the right. The enzymes have secondary, tertiary, and quaternary structures optimized to perform best at the body temperature of about 37 oC. Denaturation inactivates the enzymes. It is reversible if the temperature is slightly above body temperature, but enzymes are denatured beyond repair at a much higher temperature.
 pH
pH
The enzymes work best at the pH of the tissue or organ where they work. This is because the structure of enzymes is optimized for the pH at which they usually function. An increase or a decrease in pH from the optimum value disrupts the secondary, tertiary, and quaternary structures and causes a reduction in enzyme activity. The effect of a slight change in pH is reversible, but a significant difference in pH denatures the enzymes permanently.
The optimum pH for most enzymes is around the physiological pH of 7.4. Still, some enzymes have different optimum pH, depending on the pH in their natural environment, as illustrated in the figure on the left. For example, the optimum pH of the starch-splitting amylase is pH 7-7.5 in the mouth. Pepsin breaks down proteins at around pH 2 in the stomach. Trypsin breaks down proteins at pH 8 in the intestine.
Enzyme inhibition
Inhibitors are substances that make enzyme lose their catalytic activity. Inhibitors prevent the substrate from binding with the active site of the substrate. The inhibition may be reversible or irreversible. There are two subclasses of reversible inhibitors; competitive inhibitors and noncompetitive inhibitors.
Competitive inhibitor
The competitive inhibitor has a shape similar to the enzyme's natural substrate. So, they compete with the substrate for the active site but do not react, as illustrated in Figure \(\PageIndex{8}\). Since there is competition between the inhibitor and the substrate for the active site, increasing the concentration of the substrate wins by outnumbering the inhibitor in the completion, and the enzyme regains its activity.

Noncompetitive inhibitor
A noncompetitive inhibitor does not have a shape similar to the substrate and does not bind to the active site. It binds with the enzyme outside the active area but changes the folding patterns of the protein such that the active site can not acquire the proper shape. So, the substrate can not fit into the active site properly, as illustrated in Figure \(\PageIndex{8}\).
Examples of noncompetitive inhibitors are toxic metals, like \(\ce{Pb^{2+}}\), \(\ce{Hg^{2+}}\), and \(\ce{Ag^{+}}\). Since the inhibitor does not compete with the substrate, increasing the substrate concentration does not recover the enzyme activity. However, some chemical agents can remove the inhibitor from the enzyme, and the catalytic activity can be recovered.
Irreversible inhibitor
 Irreversible inhibitors bind with enzymes and destroy their catalytic activity permanently. They usually bind with enzymes by covalent bonds that are not easily broken. Irreversible inhibitors include antibiotics, insecticides, and never gases. For example, penicillin is an irreversible inhibitor of an enzyme needed to form cell walls in bacteria. The bacteria without a complete cell wall can not survive. It does not affect the cell membranes of humans. Similarly, a diisopropyl fluorophosphate (DIPF) which is an organophosphate insecticide, binds with the \(\ce{-OH}\) of a serine residue in the enzyme acetylcholinesterase, as shown in the figure on the right. The enzyme-catalyzed reaction of acetylcholine, shown below, is needed for nerve impulse transmission. Inhibition of acetylcholinesterase blocks nerve impulses, causing paralysis.
Irreversible inhibitors bind with enzymes and destroy their catalytic activity permanently. They usually bind with enzymes by covalent bonds that are not easily broken. Irreversible inhibitors include antibiotics, insecticides, and never gases. For example, penicillin is an irreversible inhibitor of an enzyme needed to form cell walls in bacteria. The bacteria without a complete cell wall can not survive. It does not affect the cell membranes of humans. Similarly, a diisopropyl fluorophosphate (DIPF) which is an organophosphate insecticide, binds with the \(\ce{-OH}\) of a serine residue in the enzyme acetylcholinesterase, as shown in the figure on the right. The enzyme-catalyzed reaction of acetylcholine, shown below, is needed for nerve impulse transmission. Inhibition of acetylcholinesterase blocks nerve impulses, causing paralysis.



