7.8: Protein misfolding and denaturation
- Page ID
- 430200
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Understand misfolding of proteins and the medical problems associated with it.
- Understand the denaturation of proteins and what causes the denaturation.
Protein misfolding and associated diseases
Newly synthesized proteins fold in a specific way, i.e., into secondary, tertiary, and quaternary structures, to be able to perform their function, as illustrated in Figure \(\PageIndex{1}\). Some proteins can fold in only one way, but others can fold in multiple ways. There are proteins in the cell, called chaperones, that help newly formed proteins to fold in the way needed for their function.

Sometimes normal proteins misfold and become pathological. Often, these proteins are in soluble \(\alpha\) helix forms that re-assembles into \(\beta\)-pleated sheet forms that are sticky and aggregate into plaques or amyloid structures, as illustrated in Figure \(\PageIndex{2}\), with the example of plaque formation in Alzheimer's affected brain.

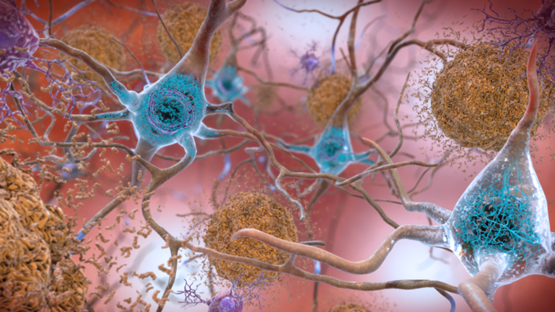
Prions are small proteins found in nerve tissue. Their exact functions are unknown, but when they misfold, they can cause more normal proteins to misfold. This protein misfolding is related to diseases such as mad cow disease in cows, Creutzfeldt–Jakob disease in humans, Alzheimer's disease, and familial amyloid cardiomyopathy or polyneuropathy, as well as intracellular aggregation diseases such as Huntington's and Parkinson's disease.
Denaturation of proteins
Proteins usually keep their primary, secondary, tertiary, and quaternary structures under physiological conditions. However, some physical processes and chemical agents can beak the interactions, i.e., hydrogen bonding, disulfide linkages, salt bridges, and hydrophobic interactions.
Loss of the secondary, tertiary, and quaternary structures of proteins by a physical process or a chemical agent while maintaining the primary structure almost intact is called denaturation of proteins.
Proteins unfold and become almost linear polypeptide chains upon denaturation. Denatured proteins can not perform their functions. Denaturation can be caused by heat, acids or bases, organic compounds and solvents, heavy metal ions, and agitation, as explained below.
- Heating above 50 oC disrupts hydrogen bonding and hydrophobic interactions, causing protein denaturation. For example, cooking food and sterilizing surgical instruments by autoclave treatment. In laser surgery, laser, i.e., the light of a single wavelength, is focused on a spot, causing heat that denatures proteins. Heating by laser cauterizes incisions, i.e., burns the site or the wound. It helps prevent blood loss.
- Acids and bases disrupt hydrogen bonding and salt bridges, e.g., by neutralizing some ions involved in the salt bridges.
- Organic compounds like urea disrupt hydrogen bonding by replacing them with stronger hydrogen bonding with the compound, and organic solvents disrupt hydrogen bonding and hydrophobic interactions. For example, 75% alcohol sterilizes skin by denaturing and coagulating proteins.
- Heavy metals ions like \(\ce{Pb^{2+}}\) and \(\ce{Hg^{2+}}\) attack \(\ce{-SH}\) groups making salt bridges, like \(\ce{-S^{-}Pb^{2+} ^{-}S{-}}\). Egg white or milk is an antidote to heavy metal poisoning as they precipitate the metal ions in the stomach, based on the salt bridge forming reaction. Vomiting is induced to remove the metal before the precipitate is dissolved, releasing the metal in the later parts of the digestive system.
- Agitation physically disrupts hydrogen bonding and hydrophobic interactions. Whipped cream and whipped egg white are prepared based on the agitation process.


