LAB 8 - MOLECULAR MODELS (Stereochemistry)
- Page ID
- 506285
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Stereochemistry – 3D Printing
The purpose of this experiment is to:
- Use drawings to try to determine if pairs of isomers are constitutional isomers, geometric isomers, or stereoisomers.
- Also, use the drawings to determine if the stereoisomers are enantiomers or diastereomers.
- Use printed 3D models to confirm your predictions.
INTRODUCTION
The structure of molecules can differ, even if they share the same molecular formulas. Molecules with identical formulas but different arrangements of atoms are called isomers. The way atoms are connected and their spatial positions in three dimensions often cause isomers to have very different physical, chemical, and biological properties. In this lab, you will investigate various types of isomers by creating computer models to visualize their structures. Additionally, you will build and print 3D models to help you compare the molecular structures of some isomers.
PRE-LAB QUESTIONS
Name ____________________________________
1) Define the following terms.
Constitutional isomers:
Geometric isomers:
Stereoisomers:
Enantiomers:
Diastereomers:
Meso compound:
Be sure to follow all your instructor’s instructions regarding the proper use of your 3D printer. Parts of the printer can be hot, and the mechanical moving parts can cause injury unless proper care is taken.
EQUIPMENT NEEDED
- 3D printer
- Computer with the 3D chemical structure drawing and 3D printing software installed (described below)
EXPERIMENTAL PROCEDURE
The following programs will be used for the instructions for this lab. These are all available free of charge.
- Jmol[1] to create a 3D computer model of a molecule and export it as an STL file.
- Ultimaker CURA[2] to slice the STL file and create the G-code needed by the 3D printer.
- More details on how to attain and use these programs are given in the appendices at the end of this document.
Part I – Constitutional Isomers
1. For each of the following molecular formulas, determine two constitutional isomers.
2. Sketch each of the isomers and give the correct name for the compound.
3. Use Jmol to make a 3D computer model of each of the isomers. Use File→Get MOL, enter the name of the molecule, and press OK. The window should display a 3D model of the molecule. See the appendix for more details.

4. Select Tools→2-D Editor to open a window showing a drawing of your molecule. Compare the program’s drawing with your drawing. How are they different?

5. Repeat the above procedure for each of the chemical formulas.
6. Choose one of the pairs of constitutional isomers. Export each isomer as an STL file and print it. Use the 3D printed models to compare the structures and explain why these are constitutional isomers.
|
Molecular |
Constitutional |
Constitutional |
Observations and |
|---|---|---|---|
|
C4H10 |
Name: Sketch: |
Name: Sketch: |
|
|
C3H8O |
Name: Sketch: |
Name: Sketch: |
|
|
C2H6O |
Name: Sketch: |
Name: Sketch: |
Part II –Geometric Isomers
1. What are the complete names of the two isomers of 2-butene?
2. Sketch each of these isomers.
3. Use the complete name of each of the isomers to get the MOL files into Jmol and observe the 3D models. Briefly describe the differences between the two isomers.
4. Transfer the 3D models to the 2D window and compare the drawings with yours.
5. Repeat the above exercise with 1,2-dichlorocyclohexane.
6. Choose either the pair of 2-butene isomers or 1,2-dichlorocyclohexane isomers, export the STL files, and make 3D prints of the pair. What are your observations after looking at the 3D printed models?
|
Molecule |
Geometric |
Geometric |
Observations and |
|
2-butene Molecular Formula: |
Complete Name: Sketch: |
Complete Name: Sketch: |
|
|
1,2-dichlorocyclohexane Molecular Formula: |
Complete Name: Sketch: |
Complete Name: Sketch: |
Part III –Stereoisomers
A – Molecules with one chiral center.
Alanine is one of the amino acids used to build proteins, which are essential for biological organisms.
1. The molecular structure of alanine has a central carbon connected to hydrogen and three different groups, an amine group, a carboxylic acid group, and a methyl group. Sketch the two isomers of alanine indicating the 3D structure and give the complete name for each of the isomers.
2. Use the complete name for each to load the structures into Jmol using Get MOL.
3. Transfer each of the 3D models to the 2D drawing window and compare the drawing with your sketches.
4. Export each of the isomers as an STL file and 3D print them. Compare the models of the two isomers and record your observations.
|
Molecule |
Stereoisomer #1 |
Stereoisomer #2 |
Observations and |
|
alanine Molecular Formula: |
Complete Name: Sketch: |
Complete Name: Sketch: |
B – Molecules with more than one chiral center.
1. 2,3-butanediol has two chiral centers and will have three isomers. Why aren’t there four isomers?
2. Sketch each of the three isomers and give their complete names.
3. Use the complete name for each to load the structures into Jmol using Get MOL.
4. Transfer each of the 3D models to the 2D drawing window and compare the drawing with your sketches.
5. Export each of the isomers as an STL file and 3D print them. Compare the models of the three isomers and record your observations.
2,3-butanediol Molecular Formula:
Complete Name |
Sketch |
Observations and
|
|
|
Stereoisomer #1 |
|||
|
Stereoisomer #2 |
|||
|
Stereoisomer #3 |
POST-LAB QUESTIONS
Determine whether the following pairs of compounds are the same, different, resonance structures, constitutional isomers, enantiomers, or diastereomers
1.


2.

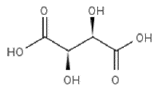
3.


4.


5.


6.

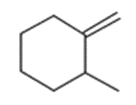
7.


Appendix A – Creating a 3D computer model.
1. Open Jmol by clicking on the Jmol.jar file. This will open the 3D window of the application.

2. Select Get MOL from the File menu. You can enter each of the molecules in the Data Table as a formula (e.g. CF4) or by name (e.g. tetrafluoromethane). Sometimes the formula will not work. If you have difficulties, ask your instructor for assistance.
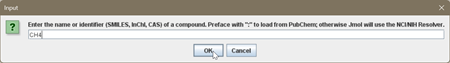
3. When you press enter or click on OK, a 3D model of the molecule will be displayed in the window.
Appendix B – Exporting and printing the 3D model
1. Export an STL file.
a. Right click on the Jmol display window and open successive menus to select File►Export►Export STL 3D model.

b. Choose a folder and give your file a name with the .stl extension.

2. Slice the STL file in CURA.
a. Open CURA then click on the open file icon.

b. Browse to where you saved the STL file and open it.
c. The model will be displayed on the printing surface. Right-click on the printing surface and select “Arrange all models” to move your model to the center of the surface.

d. You can do various manipulations to the 3D model, such as scaling it and tilting it, but usually, the default will work well. Ask your instructor for help if you are unsure.
e. Make sure that you include supports for your print. The Tree type supports work well with these models and usually selecting “Touching Buildplate” will be sufficient. If you have trouble with the print, try “Everywhere” for your support placement and as your instructor for assistance.

f. Click on the SLICE button. When the slicing is done, the program will display the amount of time needed to print the model. Click on the “Preview” button to see what the print and supports will look like. Get your instructor's approval before continuing.

g. Export the G-code by clicking on the Save to Disk button.
h. Copy the .G-code file that you just saved to an appropriate removable card as per your instructor.
3. Print the model.
a. Insert the memory card in your printer and use the printer’s control knobs to select your model. The printer will start warming up the nozzle and should begin printing within a few minutes.
b. Once the printing is complete, carefully remove the model and its supports from the printing surface.
c. The supports should be carefully removed using needle-nose pliers or other tools provided by your instructor.
Appendix C – Required Programs
· A program to create and save a stereolithography file, known as an STL file. This is a 3D solid model that can be “sliced” and printed.
· Jmol (requires java): https://sourceforge.net/projects/jmol/files/Jmol/. The download consists of a zip file containing all the files needed to run the program. Simply extract the files to a folder of your choice. The program is started by clicking the Jmol.jar file.
· Note: If Java is not available on your computer, an online version can be accessed on the Hack-a-mol website (https://chemapps.stolaf.edu/jmol/jsmol/hackamol.htm).
· A slicer program to prepare the STL file for printing and generate the G-code that is needed by the printer to make the print.
· Your printer may come with a slicer program, in which case it should be used.
· One of the best is Ultimaker CURA (https://ultimaker.com/software/ultimaker-cura/) The program must be installed on your computer, so some administrative rights are required.
· There are also several free sites that can generate G-code from an STL file online. An example is Creality Cloud https://www.crealitycloud.com/blog/tutorials/how-to-convert-stl-to-gcode. Many others can be found by searching the web.
[1] Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/


