3.1: Metal-Ligand Interactions
- Page ID
- 192538
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Hard-Soft Acid-Base Theory (HSAB)
HSAB Theory categorizes metals (acids) and ligands (bases) into different categories based on their “Hard” and “Soft” character. Character is somewhat dependent on the charge density (charge/radius ratio) of the group, but is also dependent on the type of interaction occurring. In general, Hard atoms interact with more electrostatic character (+/- attraction) and soft interactions result in bonds with more covalent character. Get out your crayons! Use the information in the table below to color-code the periodic table according to hard, borderline, and soft character.
|
|
Hard |
Borderline |
Soft |
|---|---|---|---|
|
Acids |
H+, Li+, Na+, K+, Mg2+, |
Fe2+, Co2+, Ni2+, Cu2+, |
Cu+, Ag+, Au+, Cd2+, |
|
Bases |
F⁻, Cl⁻, H2O, OH⁻, O2⁻, |
Br⁻, NO2⁻, SO32⁻, SCN⁻, C6H5N |
H- , R- , I⁻, SCN⁻, H2S, |
*In the case of bases that can bind through different types of atoms, the underlined element is the site of connectivity to which the classification refers.
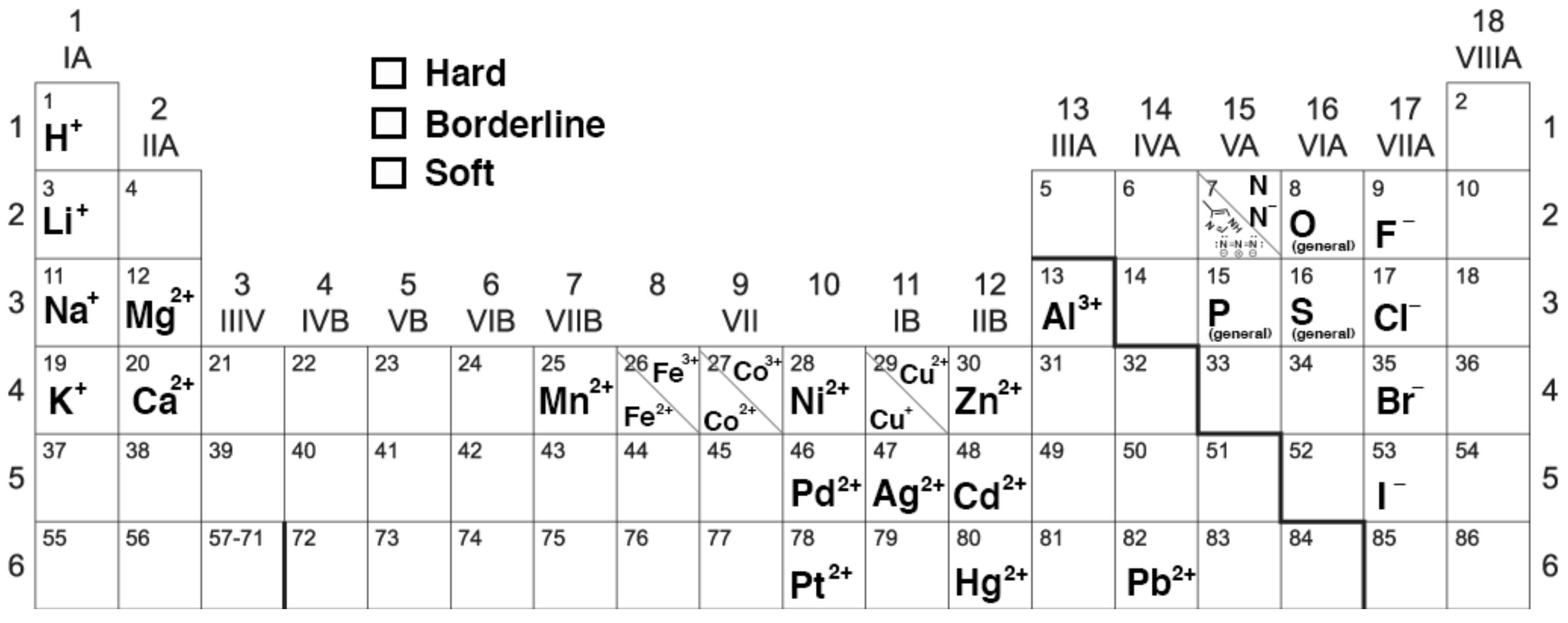
1. What trends do you notice?
(a) In general, where are acids positioned on the periodic table? ...Hard acids? ...Soft acids?
(b) In general, where are bases positioned on the periodic table? ...Hard bases? ...Soft bases?
(c) In general, how do position on the table and ionic charge contribute to hard, borderline, and soft character?
2. Consider K and Cu.
(a) Which atom has a larger atomic radius? Which has a larger radius for its +1 ion?
(b) Based on charge density alone, which ion would you expect to be softer: K+ or Cu+? Which of these two ions is actually softer based on the chart above?
(c) How does this fit into the generalizations about size and HSAB character? What other factor(s) must be considered to explain this discrepancy?
3. Come up with additional generalizations you could use to predict HSAB character of a ligand (base). Use the prompts below to guide you.
(a) Characterizing a base from the type of donor atom (O,N,S)...
Hint: It may help to highlight the donor atom for each base listed in Table \(\PageIndex{1}\).
(b) How and when does resonance matter?
(c) Alkali metals...
(d) Metals of row 4 with +2 charge? ...their corresponding +3 ions? ...their corresponding +1 ions?
4. What do the the nitrogen donor atoms from imidazole, pyridine, and azide have in common? Why are these N’s borderline while all other N’s are hard?
Proteins as ligands
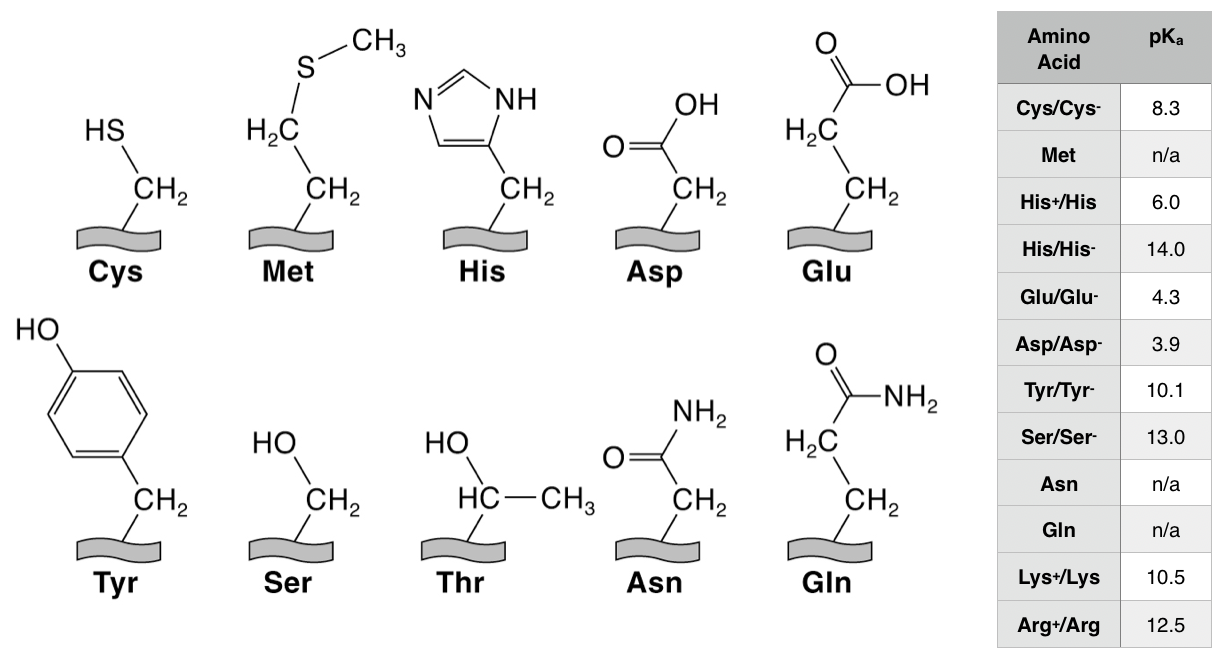
5. The amino acids that typically coordinate to metal ions are shown below. Label each of these side chain groups as hard, borderline, or soft bases.
6. The amino acids above are shown in their neutral form. Which of them will actually appear in their negatively charged conjugate base form at biological pH (7.4)? **pKa’s are given in the table.
7. Use what you know about HSAB theory to match the following metal ions to the picture of ther appropriate binding sites. Explain/defend your choices.
a) Fe2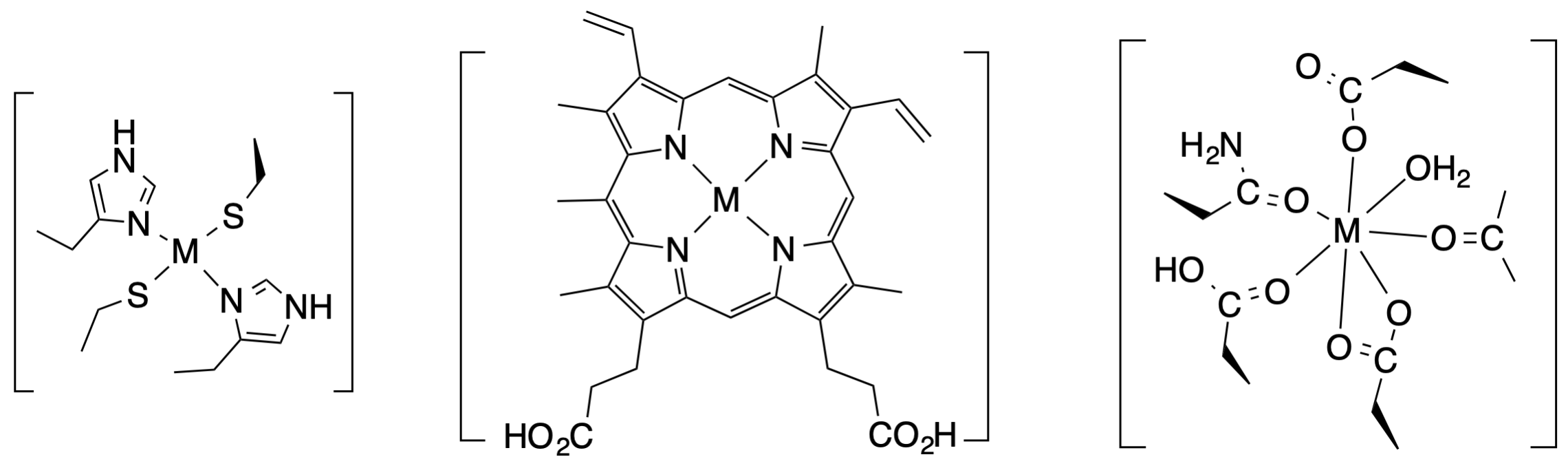 +/Fe3+ binding site (binds BOTH oxidation states)
+/Fe3+ binding site (binds BOTH oxidation states)
b) Zn2+ binding site
c) Ca2+ binding site
Application of HSAB in biology: Insight into human diseases and toxicity.1
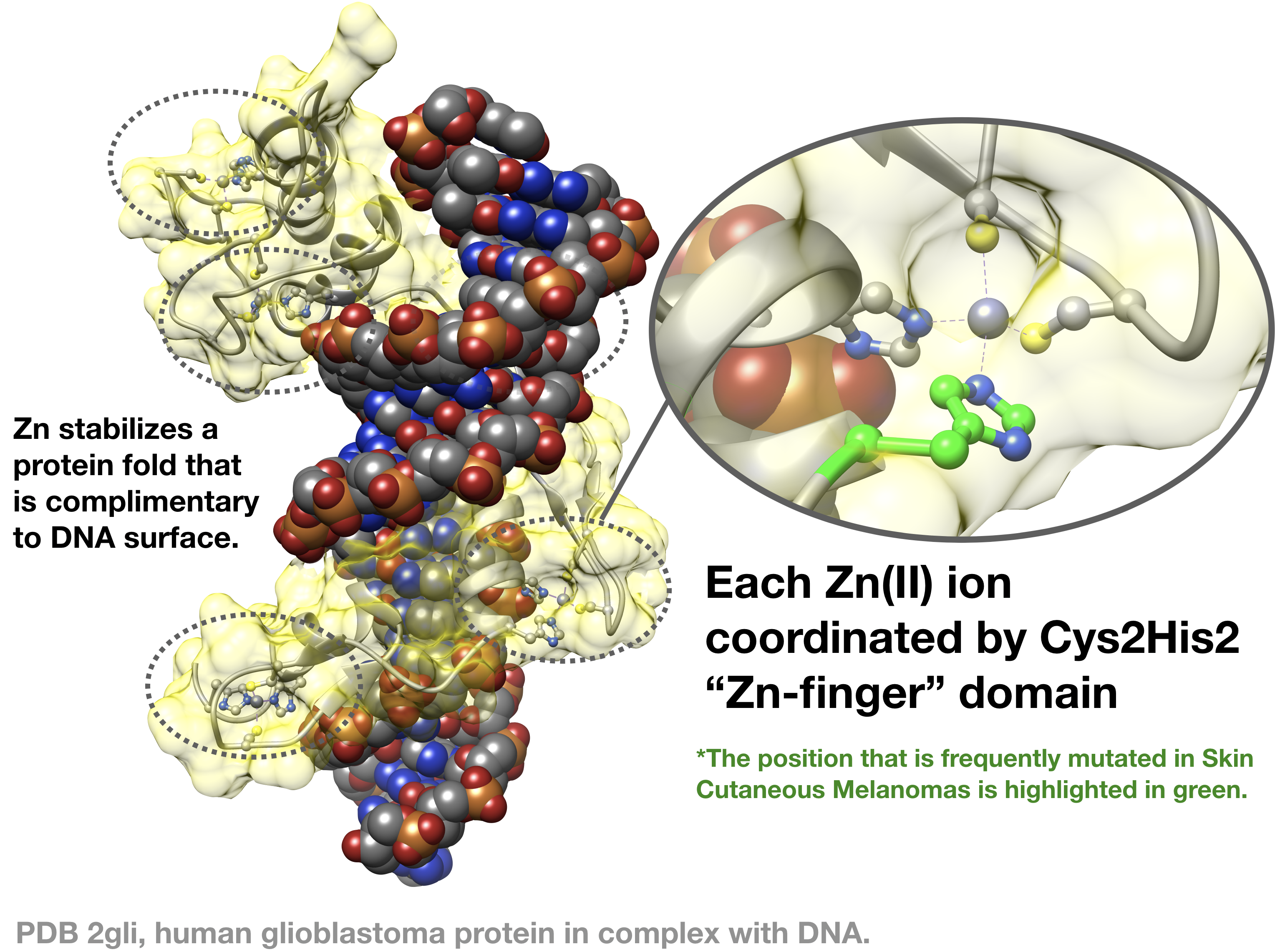 Many proteins use Zn2+ as a structural scaffold for protein folding. These proteins employ "zinc finger" domains that bind to Zn through combinations of cysteine (Cys) and Histidine (His) side chains. Zinc binding to these domains stabilizes a specific protein fold that is critical to the protein's function. Over half of transcription factors have zinc finger domains that allow the proteins to recognise and bind to DNA. Many other proteins also have zinc-finger domains and these domains are important for a variety of functions including DNA-binding, RNA-binding, protein-protein interactions, and catalysis.
Many proteins use Zn2+ as a structural scaffold for protein folding. These proteins employ "zinc finger" domains that bind to Zn through combinations of cysteine (Cys) and Histidine (His) side chains. Zinc binding to these domains stabilizes a specific protein fold that is critical to the protein's function. Over half of transcription factors have zinc finger domains that allow the proteins to recognise and bind to DNA. Many other proteins also have zinc-finger domains and these domains are important for a variety of functions including DNA-binding, RNA-binding, protein-protein interactions, and catalysis.
The human glioblastoma protein (GLI) family is a class of transcription factors that demonstrate sequence-specific DNA binding through "classic" Cys2His2 Zn-binding motifs. These proteins are essential during embryonic development. Dysfunction of these proteins are linked closely to glioblastoma and other aggressive cancers. The zinc finger domain of GLI1 is shown in this image (PDB 2gli),2 where the protein surface is highlighted in yellow. GLI1 possesses five Cys2His2 zinc finger domains. When Zn is not bound to the protein, it does not have the appropriate structure to bind to recognize DNA.
The size and structure of the Zn2+ ion is critical to provide the correct protein fold for DNA binding. Thus, zinc fingers must bind specifically to Zn (as opposed to other biologically available metal ions). The zinc finger domains use principles of inorganic chemistry to control metal ion selectivity. While other metal ions, like Co2+, Ni2+, Fe2+, and Mn2+, can bind in the zinc finger sites, Zn2+ binds 1,000 to 100,000 times more strongly than any of these other metals.
8. Apply HSAB to explain why there is such a strong selectivity for Zn2+ binding in a zinc finger site as opposed to other metals, including Co2+, Ni2+, Fe2+, and Mn2+.
9. Cadmium (Cd2+) and lead (Pb2+) are toxic metal ions that bind more strongly than Zn2+ to zinc binding sites in proteins. Why do these toxic metals compete well with Zn2+, while other metals like Co2+, Ni2+, Fe2+, and Mn2+ don’t?
10. If a toxic metal, like Cd2+ would bind to the Zn2+ binding site, how and why might this affect the protein’s ability to bind to DNA?
11. It was recently discovered that mutations in zinc finger domains may be related to some cancers. For example in "classic" Cys2His2 zinc finger domains, one of the His is frequently mutated to a tyrosine (Tyr) in melanomas.3 How might this mutation affect binding to Zn2+, and how might this affect the ability of the protein to recognize its DNA sequence? (See figure above for example illustration) Explain your reasoning.
Chelate Effect
12. Describe the following in your own words:
Chelator:
Monodentate Ligand:
Bidentate Ligand:
Multidentate Ligand:
The reactions below are a classic illustration of the chelate effect:
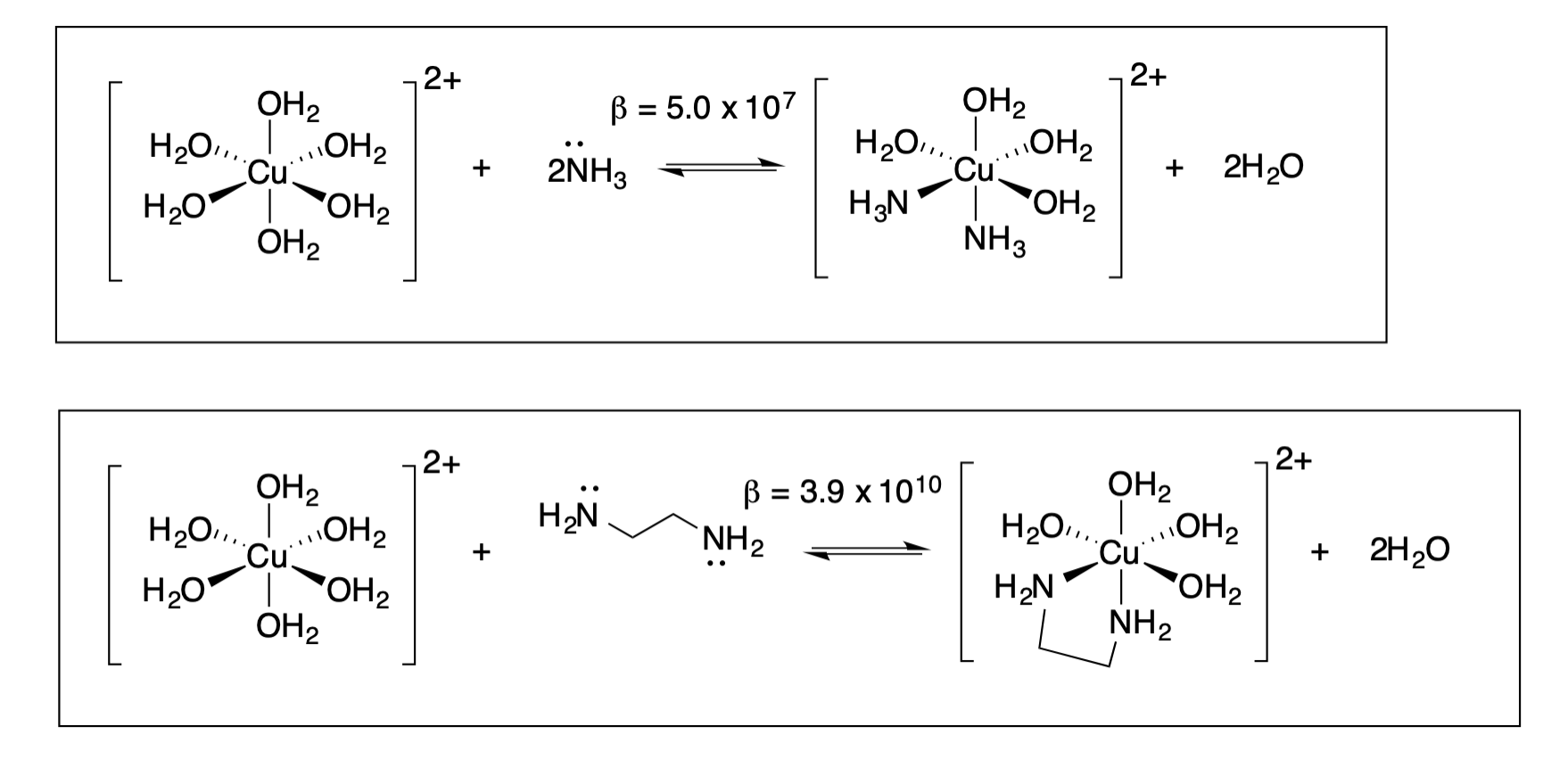
13. What kind of ligand is ammonia (NH3)? What is the donor atom(s)?
14. What kind of ligand is ethlyenediammine (NH2CH2CH2NH2)? What is the donor atom(s)?
15. The Chelate effect is an entropic effect. Increase in entropy (disorder) is favorable. Apply the entropy argument to explain why the second reaction, with a chelator, is more favorable than the first reaction.
Macrocycle Effect / Pre-organized Binding Sites
The chelate effect is an entropic benefit that extends to all polydentate systems, and the greater the number of donor atoms in one chelate, the greater the stability of the complex. The increased stability of a chelate is a result of the greater entropic advantage of displaying mono dentate ligands.
There is an additional entropic benefit observed in both macrocyclic systems in some protein-metal binding sites. For example, the ligand phthalocyanin gives complexes with metal ions that are more stable than would be expected from the chelate effect alone. The increased stability of metal complexes with macrocycles is called the “macrocycle effect” and is due to both the entropic benefit of the chelate effect combined with pre-organization of the donor atoms.
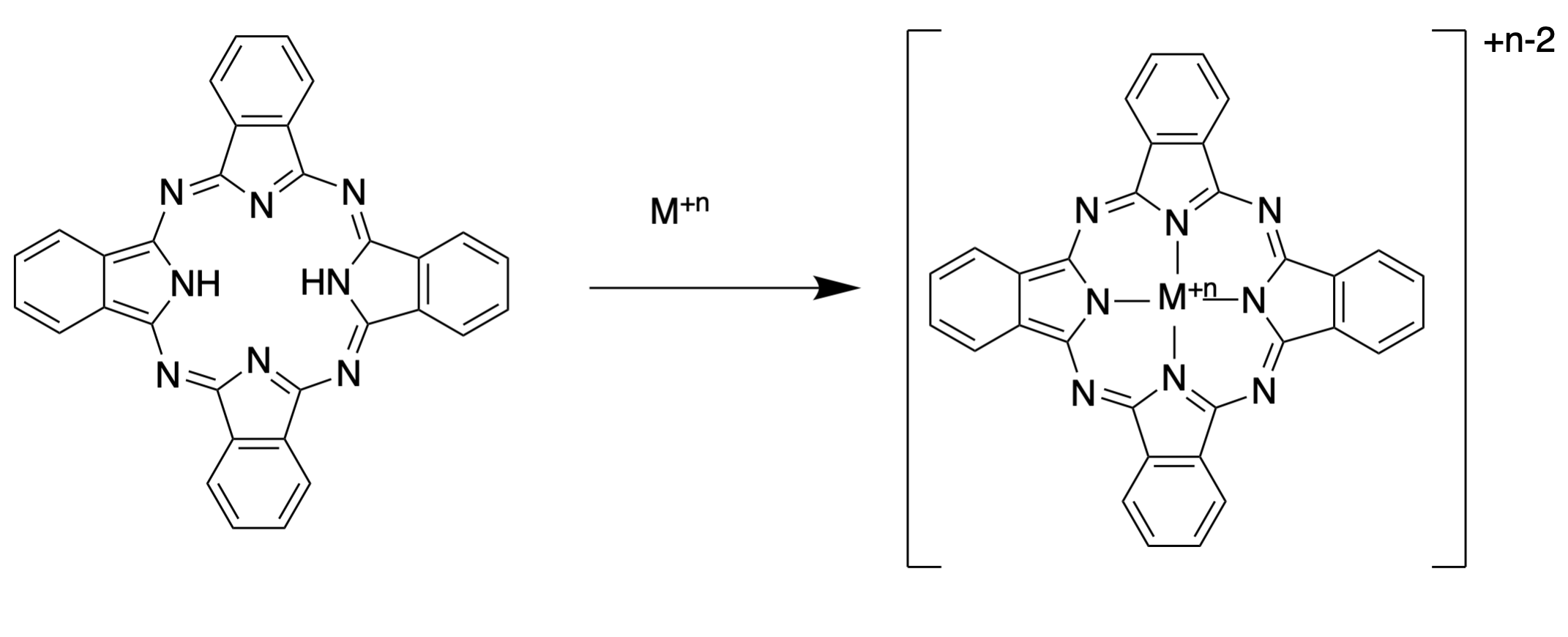
Like macrocycles, the metal binding sites in proteins are often more stable than would be expected based on the chelate effect alone. The phenomenon of preorganization extends to protein binding sites when the binding site’s donor atoms are pre-organize in close proximity by protein folding.
**Many metal binding sites in biology are multidentate chelators and/or are effected by preorganization.
16. Consider the topic you chose for your My Molecule Project. Explain how the following factors contributing to your protein/molecule being able to select and retain the "correct" metal ion?
a) HSAB (This is both a selectivity and a stability factor. Consider the atoms actually binding to the metal)
b) Chelate Effect
c) Macrocycle/Pre-organization (this can also be related to size of the metal ion!)
Sources
- Inspired by Zinc Finger Proteins: A Bioinorganic Example of LFSE and HSAB, a problem set created by Elizabeth R. Jamieson, Smith College (ejamieso@smith.edu) and posted on VIPEr (www.ionicviper.org) on June 24, 2013. Copyright Elizabeth R. Jamieson 2013. This work is licensed under the Creative Commons Attribution-NonCommerical-ShareAlike 3.0 Unported License. To view a copy of this license visit http://creativecommons.org/about/license/.
- Pavletich, N. P.; Pabo, C. O., Crystal Structure of a Five-Finger Gli-DNA Complex: New Perspectives on Zinc Fingers. Science 1993, 261 (5129), 1701-7.
-
Munro, D.; Ghersi, D.; Singh, M., Two Critical Positions in Zinc Finger Domains Are Heavily Mutated in Three Human Cancer Types. PLOS Computational Biology 2018, 14 (6), e1006290.

