2: Introduction to Metal-Ligand Interactions
- Page ID
- 192303
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)What is a metal-ligand interaction?
Metal-Ligand interactions are just Lewis Acid/Base reactions!!!
The bonding between metals and ligands can occur on a spectrum of covalence and strength. Some metal-ligand bonds are similar to ionic interactions, while others are essentially covalent. Bonds between metals and ligands are commonly referred to as "coordinate covalent" bonds.
Regardless of the nature of the metal-ligand interaction, when a metal ion interacts with a ligand it is fundamentally an acid/base reaction! Metal-ligand interactions also fit right into what you learned about nucleophiles and electrophiles from organic chemistry!
1. Define the following:
a) Lewis Acid / Electrophile:
b) Lewis Base / Nucleophile:
Same type of reaction, different formalisms
Formalisms are different in organic chemistry and inorganic chemistry, particularly where formal charges are placed and where electrons are placed in our drawings.
Label the Lewis acid, electrophile, Lewis base, and Nucleophile on the reactants side in each box:
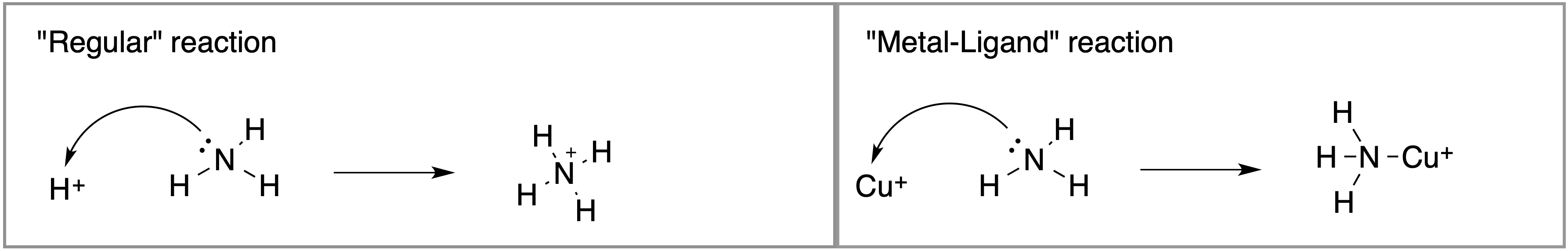
2. Compare the two reactions shown above.
a) How are the two reactions above similar?
b) How are they different?
3. Look at the first, “regular”, reaction above:
Notice that formal charges change from reactants to product. What does this say about change in “ownership” of electrons in the new N-H bond?
4. Look at the the second, “metal-ligand”, reaction above:
Notice that formal charges do not change from reactant to product. What does this indicate about the change of “ownership” of electrons in the N-Cu bond?
5. **Please note that formal charges are not “real”, and they can be deceiving! Formal charges are very poor indicators of where electron density actually exists! To illustrate this, draw electron dipole arrows on a Lewis Structure of NH4+. Where is the formal positive charge; on N or on H's?? Which atoms are electron deficient according to the dipole moment arrows?
The shapes of metal coordination structures
One of the reasons that metal complexes are so useful in biology is due to their broad repertoire of available structures (shapes). If we only had carbon, biology would be limited to just a few shapes (linear, bent, trigonal plane, and tetrahedron) and bond angles (180, 120, 109.5); but with metal ions, so much more is possible.
6. This is a scavenger hunt: Please go to the Metal Ions in Biology chapter in the Readings section of this course and find at least three examples of metal coordination structures that would not be possible from carbon. Please name the biomolecule, state what the role of this biomolecule is, and sketch the coordination geometries (by hand) that you find.
a) Example 1: Name____________________________ Function _________________________
b)Example 2: Name____________________________ Function _________________________
c) Example 3: Name____________________________ Function _________________________
Biomolecules as ligands
Any nucleophilic atom (or functional group) on a biomolecule can act as a ligand! This includes the nucleophilic atoms on proteins, DNA, carbohydrates, fatty acids, and small molecules!
Example: Amino Acid Side Chains
Let's take a look at how the nucleophilic groups in a polypeptide might interact with a metal ion:
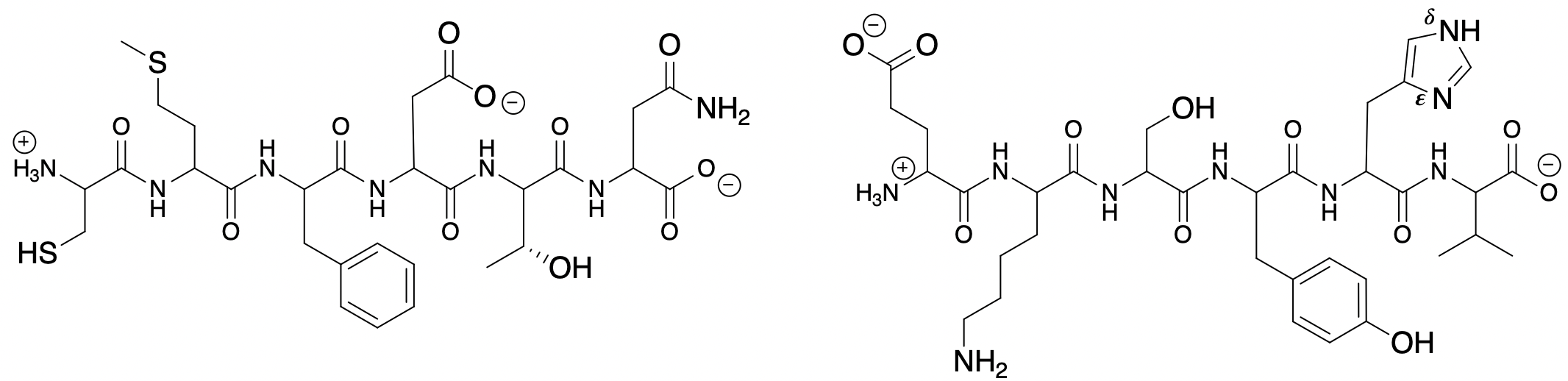
Above are the structures of two polypeptides. The atoms of amino acid side chains are shown in larger font than those of the peptide backbone. The side chains and backbone heteroatoms within a polypeptide are “ligands” that typically coordinate to metal ions. If something is acting as a ligand for a metal ion, it must be acting as a Lewis Base.
7. Identify the amino acids. For each one, decide if its side chain can coordinate to a metal ion. Then, highlight the atom on each amino acid that is the most likely to bind directly bind to a metal ion. **First, you’ll need to recall what ALL Lewis bases have in common, and how to determine relative base strengths (You learned this in Organic Chemistry).
8. When the amide side chains of asparagine (N, Asn) and glutamine (E, Gln) bind to metals, why do they bind through their O atom, and not N? Draw resonance structures in your response.
Transition metals alter the pKa's of ligand protons
Transition metal ions (but not alkali metal ions) act as Lewis acids in metal-ligand interactions. And also, the metal-ligand complex itself can act as a Bronsted acid (giving off a proton). When a ligand has an acidic proton, interactions with a metal ion will make that acidic proton more acidic. In other words, interaction with the metal ion can cause a decrease in the ligand's pKa.
Example: Water meets a transition metal ion1
In biology, the solvent is water. Water is also a ligand that binds to metal ions, according to the equation shown below.
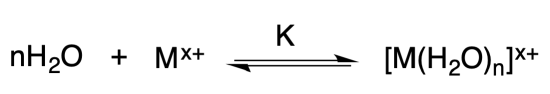
There is a LOT of water around in biological systems. Consequently, the reaction shown above is shifted far to the right in aqueous solution. Consequently, when metals are dissolved in aqueous solution, they exist as a complex with water. This is generally true unless there are other ligands present that have stronger affinity for the metal ion.
Consider the two equilibrium reactions shown below. The autoionization of water is shown in the box on the right. This reaction has a very small equilibrium constant (KW = 10-14) and thus the pKa for dissociation of a proton from water is pKa = 14. On the left is the dissociation of a proton from a water molecule bound to a Zn2+ ion. The bond between water and Zn is considered to be covalent.
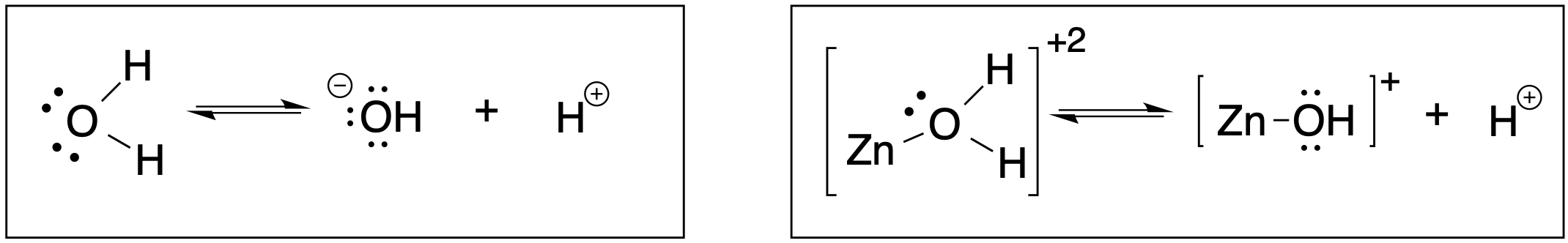
9. Predict which reaction above is more favorable. In other words, in which case is water a better acid; when water is "free", or when it is bound to a transition metal ion like Zn2+? Defend your prediction.1
10. Water has a pKa of 14. When water is bound to a Zn2+ ion in aqueous solution, its pKa is lowered to 10. Calculate the pH of pure water and the pH of a solution of 0.1 M ZnSO4. **Hint: You will need the reactions above and you will need to recall the definitions of Ka and pKa to complete this task!1
11. Transition metals dramatically decrease solution pH compared to pure water. On the other hand, alkali metals do not significantly affect pH.
a) Based on this information, what can you conclude about the relative covalent nature of water’s interactions with transition metals compared to alkali metals?
b) Rationalize this based on the element's position on the periodic table and their consequent electron configurations.
12. When a transition metal binds to a Cys side chain, it usually binds to the thiolate anion form of this side chain. Explain why the Cys loses its proton when a metal ion binds. Use drawings to describe what happens to the pKa of the thiol hydrogen.
Biological application for metals that alter ligand pKa: Hydrolytic enzymes2
Above you learned how metals can act as Lewis Acids to increase the acidity of a bound water molecule (or other ligand). This modulation of water’s pKa can be really useful to catalyze reactions in which hydroxide ion is a reactant. The enzyme carbonic anhydrase (CA) is a prime example of how biology uses this property to catalyze an important biological acid/base reaction. CA is the enzyme that converts CO2 into bicarbonate (HCO3-) very quickly. This is important in the blood to get to CO2 to the lungs to be exhaled. Conversion of CO2 to bicarbonate is also important in production of the aqueous fluid of eye and other secretions; and CA is an important drug target for treatment of glaucoma.
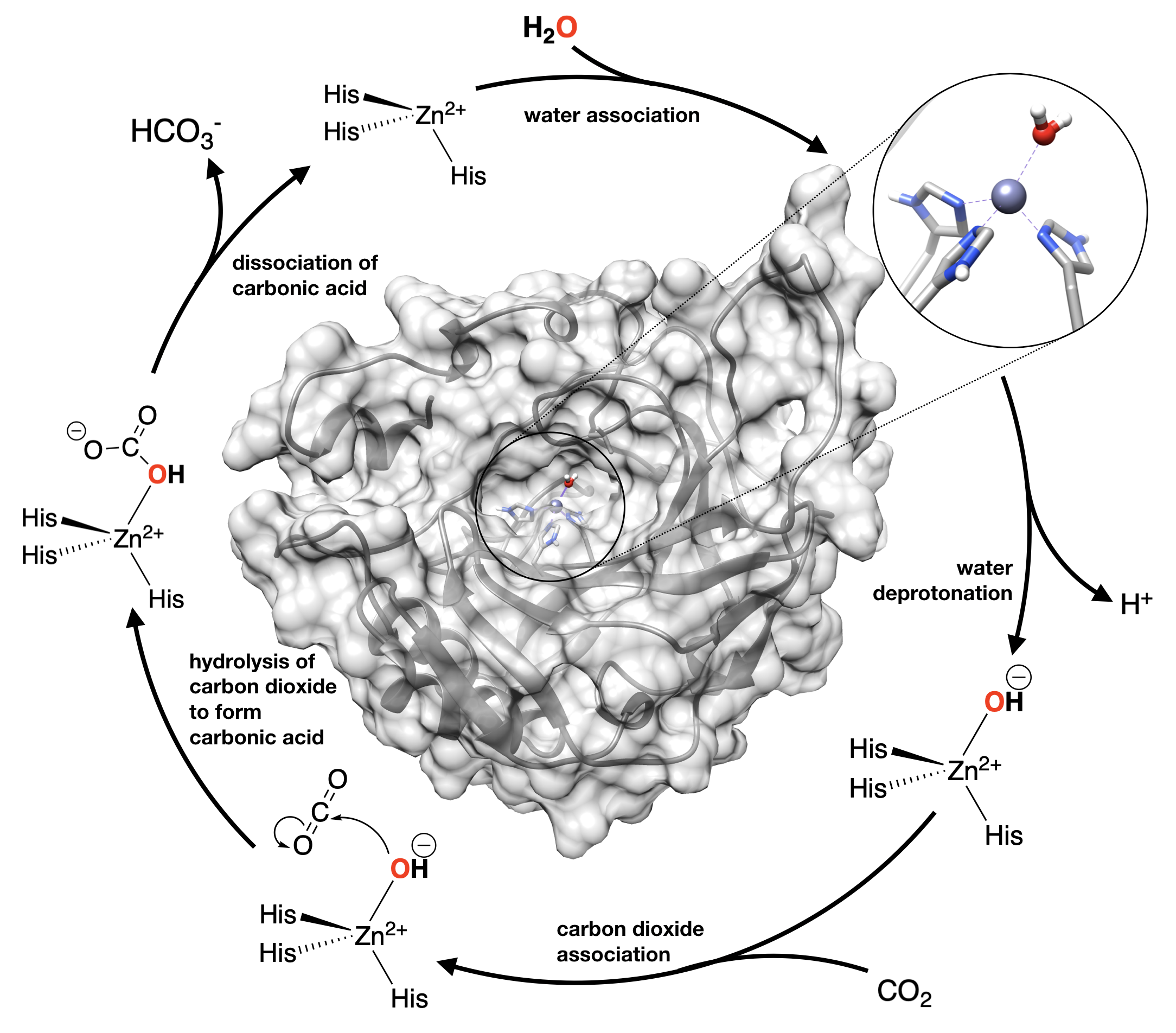
The figure shows the structure of human carbonic anhydrase IV (from PDB 1znc), with a blow up of the Zn active site where Zn is bound to 3 histidine side chains (His). The mechanism for this enzyme is shown around structure of the protein. As you saw earlier, binding water to a metal ion can change its pKa, and make its protons more acidic by stabilizing the hydroxide ion. When hydroxide is needed to carry out hydrolysis reactions, acidic metal ions, like Zn2+ are often used to stabilize the hydroxide nucleophile in order to catalyze the reaction (lower activation energy). The pKa of “free” water is 14. For water in [Zn(H2O)6]2+ in aqueous solution, the pKa is 10. In the active site of carbonic anhydrase, the pKa of water lowered even more, to 7, by the protein environment around the Zn active site.
13. Which step of the mechanism would be nearly impossible in pure water?
14. The CA active site pocket is hydrophobic: consider how well charge would be stabilized in aqueous solvent compared to in a hydrophobic pocket. Explain how a hydrophobic metal binding site would decrease the pKa of water bound to Zn in CA compared to that of water bound to Zn in “bulk” solution.
Sources
1. Inspired by or taken directly from Metals in Acid Base Chemistry, an in-class activity created by Sheila Smith, University of Michigan- Dearborn (sheilars@umd.umich.edu) and posted on VIPEr (www.ionicviper.org) on October 17, 2009. Copyright Sheila Smith 2009. This work is licensed under the Creative Commons Attribution Non-commercial Share Alike License. To view a copy of this license visit http://creativecommons.org/about/license/.
2. From Metals in Biological Systems - Who? How? and Why?, a 5 slides about learning object created by Adam R. Johnson, Harvey Mudd College (adam_johnson@hmc.edu), Hilary Eppley, DePauw Univeristy (), and Sheila Smith, University of Michigan- Dearborn (sheilars@umd.umich.edu) and posted on VIPEr (www.ionicviper.org) on January 20, 2010. Copyright Sheila Smith 2009. This work is licensed under the Creative Commons Attribution Non-commercial Share Alike License. To view a copy of this license visit http://creativecommons.org/about/license/.

