Biochemistry: Chloramine Processing Protocols
- Page ID
- 77871
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Objectives:
- Generate chloramine in deionized water to produce similar byproducts and achieve a standard of 4.0 ppm monochloramine concentration equivalent to upper range concentration of local tap water(U.S standard chloramine water treatment adapted from water municipality method)
- Evaluate six treatment methods (including controls) to effectively breakdown chloramine in water in ways that will not adversely effect fish, plants or consumers. (e.g. Common aquarium methods using sulfides would cause high toxicity in consumers of fish and plant crop.) Biomagnification must be considered as there are is potential for toxins to increase exponentially going from fish to plant to consumer.
- Measure chloramine concentration and rate of breakdown by comparing Total chlorine vs. Free chlorine in six to indicate Chloramine break down by choosing the appropriate detecting test.
- After breakdown of chloramine the elimination of free chloride via off gasing will be paramount to inhibiting formation of insoluble oxides in the presence of organic ions (nutrients Fe2+….).
Methods:
1. Generating Chloramine Water with a total of 6 barrels
initial equation: NH3 + Cl2 --> NH2Cl (pH: 8.4)
1 : 4 (similar ratio to tap water)
The starting chloramine concentration has to be 4 ppm = 4 mg/L = Molarity of 7.77 x10-5 M. Therefore, HOCl(s) + NH3(aq) will be used to make monochloramine(NH2Cl) at pH of 8.4. The byproducts of this reaction will include trihalomethanes (eg. CHCl3...) and Nitrogen trichlorides (NCl3) that will be considered as interference factors for measuring the monochloramine(These are the same byproducts present in tap water). An additional DPD colorametric test step of using thioactamide may be used to diffreniate the monochloramine concentration from nitrogen containing chlorides.
*Testing at pH 4.0 may will require H2SO4 for lowering pH Buffer.
2. Monochloramine water will be tested standards will be tested for proper concentration. All colorimetric testing preparations should be done complete prior to the treatment. (e.g. Standardized the solution with KI in cuvettes before adding samples)
3. TREATMENTS
The monochloramine water generated will be in six separate barrels. One barrel will be a control measuring any auto-decomposition and off gas over time (likely to take weeks). The other five barrels will correspond to the five treatments to process the chloramines will be performed, along with the control of a standing water. O2 from air at the surface of the water and being pumped and bubbling in the tanks should act as a catalyst for the decomposition of chloramine. down in a course of one week.
*All treatments will start with the same as the control barrel; 5 gallons of DI water with 4 mg/L (4ppm) of monochloramine (CH2Cl).
a. Sodium Ascorbate
Treatment A will using Sodium Ascorbate to process the monochloramine. (Asorbic acid/Vitamin C works through the same pathway as Sodium Ascorbate but requires 50x more concentration than Sodium Ascorbate. Since both treatments are adding acid to the system the Sodium Ascorbate is the preferred method as it will not decrease the pH as much as the Asorbic Acid.)
*pH must fall below 5 (as close to 4 as possible) for following reactions. At this pH the monochloramine will be converted to di/trichloramine.
Ascorbic acid:
C5H5O5CH2OH + HOCL → C5H3O5CH2OH + HCl + H2O
Ascorbic acid + Hypochlorous acid → Dehydroascorbic acid + Hydrochloric acid + water
Sodium Ascorbate:
C5H5O5CH2ONa + HOCL → C5H3O5CH2OH + NaCl + H2O
Sodium ascorbate + Hypochlorous acid → Dehydroascorbic acid + Sodium chloride + water
b. Nitrofiles (Bacteria):
The bacteria nitrosimonas and nitrobacter will be cultured and cured on live rocks (Fiji Rock) within the water tank to support the nitrification process;
The biochemical reaction of Nitrosomonas:
The next stage is a direct oxidation step, as follows:
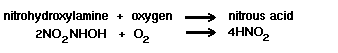
Sixty-six kilocalories of energy are liberated per gram atom of ammonia oxidized.
The biochemical reaction of Nitrobacter is a very simple reaction, involving the cytochrome system as follows:
Then the cyt.Fe+++ is regenerated by:
Eighteen kcal of energy is liberated per gram atom of nitrite oxidized.
This whole process removes electrons from a hydrated nitrite ion. The reactions of Nitrobacter are inhibited by small quantities of ammonia gas (NH3: 1.4 mg/L inhibits 99%), which can lead to a toxic buildup of nitrite, since Nitrosomonas is not inhibited from oxidizing ammonia to nitrite, in the presence of ammonia.
c. Air (negative control)
Compressed air will be bubbled through the tank to drive oxidation of monochloramine. ~This method is expected to take a week to achieve safe levels of chloramine.
d. Air + bacteria
The live rocks containing the nitrofiles will be placed in tank with compressed air to
e. Ozone (control)
An aquarium pump with an ozone filter will be used to test rate of CH2Cl clearance. (Ozone may create byproducts that may not be conducive to overall aquaponics project and cause interference with DPD testing)
f. Control
The solution will sit in a barrel to establish a baseline of how concentrations change overtime without any treatment.
Data Collection
Three methods for collection will be used: DPD Titration (Kit), Pool Test Strips, and DPD COLORIMETRIC.
*RECORD starting pH, Free Chlorine and Total Chlorine (Colorimetric will have two total chlorine values because one will use thioacetamide) with each test method.
Collect two 10mL samples from each treatment barrel every thirty minutes (first four hours) and then hourly thereafter. Use one sample for testing DPD Titration Kit and the other sample for DPD COLORIMETRIC Test.
DPD COLORIMETRIC (LINK for Calibration)
Two different solutions for total chlorine will be made, one with the thioacetamide, so that it can bind with other sources of Nitrogen containing Chlorides. The other one is without thioacetamide, so that the total chlorine will indicate CH2Cl minus the interference from NCl3, NO3-...
The pH is further adjusted to 4.0 using H2SO4 to prepare for the DPD indicator.
2. Add the DPD indicator
3. Perform a DPD test with spectrophotometer (Chemistry Lab)
4. Perform another test with chlorine strips

Tolerances of Each Detection Method
- Chlorine Test strips ($10)- precision of 0.5-2.0ppm
- DPD Titration Test kit ($55)- precision of 0.2-0.5ppm
- DPD colorimetric (Abs. 512/515nm)- precision 0.05ppm //Limitation ->Breakdown of materials before testing possible
References for Chloramine Proof of Concept:
1) DPD Colorimetric process for Chlorine
http://dnr.wi.gov/regulations/labcert/documents/training/CL2DPDfull.pdf
2) Sodium Ascorbate and Chloramine break down
https://www.fs.fed.us/t-d/pubs/html/05231301/05231301.html
3) Full Text of EPA 4500 Cl -G (Regulation and method for commercial water testing. FOCUS ON THE DPD COLORIMETRIC METHOD)
https://archive.org/stream/gov.law.apha.method.4500-cl.1992/apha.method.4500-cl.1992_djvu.txt
PDF version : https://ia600300.us.archive.org/14/i...00-cl.1992.pdf ( A more readable version, the downside is that we can't search for specific words)
4) Great website regarding the use of nitrofiles and the specific tolerancees of nitrobacter/nitrosimonas:
http://www.bioconlabs.com/nitribactfacts.html
Working Inventory
| Have: | Possible Needs: |
| Six 55 gal. drums | Aqueous ammonia (15M from stockroom) |
| Air compressor (Ryan) | Sodium Hypochlorite 1L ~$23 |
| Air hose | Chlorine Test strips ~$10 precision 0.5-2.0ppm |
| Air hose fitting (male) | DPD Titration Test kit (If only Titration) $55 precision 0.2-0.5 ppm |
| pH/Temp fluid sensors | DPD colorimetric (Abs. 512/515nm) precision 0.05ppm //Limitation->Breakdown of materials before testing possible |
| Ozone pump | Ampule or vacuum cuvette kit -$25 |
| Pipette (borrow if possible) | |
|
Potassium Permanganate Standardization solution -$20 |
|
|
Sensors Chlorine/Ammonia $196 per sensor |
|
| DI Water | |
| H2SO4 | |
| KI or selenium arsenate | |
| nano water | |
| thioacetamide | |
| ferrous ammonium std. (FAS) |

