Enzymes
- Page ID
- 56106
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Enzymes are catalysts that drive reaction rates forward. Most catalysts, but not all, are made up of amino acid chains called proteins that accelerate the rate of reactions in chemical systems. The functionality of a catalyst depends on how the proteins are folded, what they bind to, and what they react with. For protein-based catalysts, amino acid polarization lies at the core of catalytic activity.
Introduction
In chemistry, a catalyst is a chemical that drives a reaction forward. Catalysts lower the activation energy, which is the amount of energy required for reactants to form products (Figure 1). Catalysts also lower the kinetic barrier, which is needed to drive a reaction forward and backward. A certain amount of energy contained in the molecules is required when the two molecules react together to form a product. If the two molecules do not have enough energy to react, then no product is produced. By lowering the activation energy, a catalyst allows the molecules to gain sufficient energy to overcome the barrier and form products.
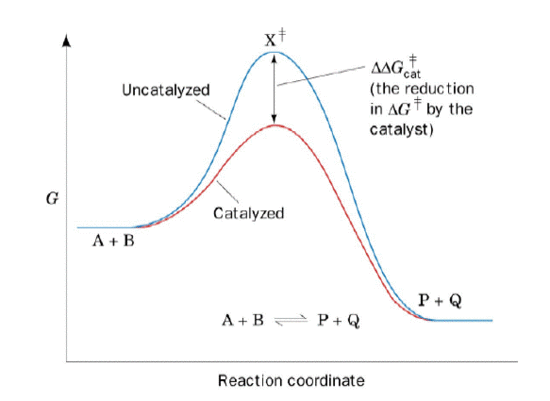 Figure 1: Compare the red curve with the blue curve. Which hill would you want to climb over? This figure shows the decrease in the activation energy and the kintetic barrier in a reaction in which there is a catalyst or enzyme (red curve). Extracted from Dr. Delmar Larsen's Lecture 22 on 5/24/10.
Figure 1: Compare the red curve with the blue curve. Which hill would you want to climb over? This figure shows the decrease in the activation energy and the kintetic barrier in a reaction in which there is a catalyst or enzyme (red curve). Extracted from Dr. Delmar Larsen's Lecture 22 on 5/24/10.
Catalysts increase the rates of the forward and backward reaction (Kf to denote the rate of the forward reaction and Kb to denote the rate of the backward reaction). Exergonic forward reactions convert reactants to products, whereas endergonic backward reactions convert products to reactants. Typically, the conversion of products to reactants requires more energy, measured in Gibbs energy. Recall that the difference in energy between the products and the reactants is measured as ΔG (Gibbs energy). It is very important to note that catalysts do not change the free energy, G, they simply affect the speed of the reaction. Catalysts are very beneficial in biological systems because they drive individual reactions forward. Our bodies are a vast combination of redox reactions. Heat may drive a reaction forward. For example, when you catch a fever, your body raises its temperature to drive reactions forward in your body, dissipating energy in the form of heat. Note that the body raises its temperature to drive reactions forward. This extra energy drives your immune system forward to get rid of the germs faster. Often, life does not want whole systems to be driven forward; instead, the biological system merely wants to produce a little extra product or a small amount of excess reactant of one reaction. It would be a waste of energy to constantly have our body hotter than needed, and we would probably die much faster. Therefore, our body uses reaction specific catalysts. These reaction specific catalysts are required to keep our body alive. In this section, we will talk about the chemistry of inorganic and organic biological catalysts, also called enzymes, and how their composition is evaluated in medicine.
Amino Acids and Polarity
What makes an amino acid polar or nonpolar? What level of polarity affects an amino acid's hydrophobicity or hydrophilic characteristics? This is the basic structure of an amino acid:
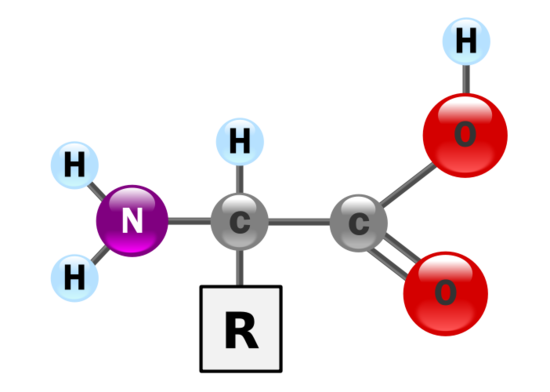
Figure 3
Notice that the basic structure always carries an amine group (NH2), and a carboxyl functional group (CO2H). The general formula for an amino acid is H2NCHRCOOH, which denotes the order in which the hydrogen and carbon atoms are bonded. The 20 amino acids have this same general structure. What makes them different is the R-linked side chain. Images of the 20 different amino acids.
Amino acids differ in their electronegativity in the R groups, causing differences in their hydrophobicity. The side chains denote whether an amino acid is:
- Basic and polar
- Acidic and polar
- Neutral non-polar
- Neutral polar
Recall that the more electronegative an R side chain is compared with its amine and carboxyl, the more polar the amino acid. In general, side chains with hydrocarbon alkyl groups (CnHn), or benzene rings are non-polar. Examples: phenylalinine, Leucine, Isoleucine The number of alkyl groups affects the polarity. The more CnHn groups, the more nonpolar.
What makes an amino acid more polar?
- Acids, amides, amines, and alcohols make an amino acid more polar.
- Acids
- Amides (the Rs can be Hydrogens in their respective prime locations)
- Amines
- Nitrogen may bond with up to four hydrogens in an organic compound to be called an amine. The most common amines are ammonia (NH3), and NH2)
- Alcohols (carbon-hydrogen chains with OH groups attached to the ends)
What makes an amino acid basic?
- An amine functional group. (Note that amino acids with amides on the side chain do NOT produce a basic amino acid)
What makes an amino acid acidic?
- Usually, carboxylic acid (COOH) groups attached to R side chains make an amino acid acidic.
Enzyme and Substrate Chemistry
Enzyme and Substrate Chemistry can be described biologically. Enzymes provide the particular substrate with an active site, which forms an enzyme-substrate complex, which is necessary for its catalyst properties and the formation of products.

In Figure 4, the particular substrate fits in the enzyme as a key fits into a lock. Extracted from Casiday and Frey Chemical-Kinetics Experiment.
The Rate of an Enzyme-Substrate reaction is proportionally related to the concentrations of both the enzyme and the substrate. As the concentration of either decreases or increases, so does the reaction rate. However, there are certain exceptions to this rule of proportionality. The reaction rate disregards or is independent of the concentration of the substrate when it is very high. Thus, the rate of the reaction for an enzyme-catalyzed reaction with a high substrate concentration follows a zero rate equation: Rate of Reaction=K. In terms of a normal proportional reaction rate to the concentration level, the rate equation is considered as first order.
CoEnzymes & CoFactors
Kinetics of Enzyme-Substrate Chemistry
Homogeneous Catalysts
Homogeneous catalysts interact with the reactants in the same phase (i.e: turning a substrate into a product at a faster rate). The homogeneous catalysts do not change their current states, unlike heterogeneous catalysts. By "states" we mean the phase state, which indicates either solid, liquid, or gas. If homogeneous catalysts are solid, then they will remain solid after the reaction is completed; the same is true for liquid and gas homogeneous catalysts. Although many biological enzymes are heterogeneous, there are some homogeneous enzymes that remain in their same state after the reaction, such as in an immunoassay (EIA).
Heterogeneous Catalysts
Heterogeneous catalysts are catalysts that speed up the rate of reactions by allowing them to occur on a solid surface. An example of a heterogeneous catalyst is a clay DNA polymer scaffolding, where the DNA's individual purines and pyrimidines link together on a clay surface to enable more secure bonding.
The Basics
Enzymes are composed of many amino acids that react with substrates in biological chemistry. Enzymes exist to drive the rates of reactions forward in our bodies. Without enzymes, products would not form quickly enough for our body to actually process the energy that we need. The basic reaction for any enzyme-substrate complex is this:
\[ \text{Step 1}\;\;\;\;\; E+S \rightleftharpoons ES \]
The enzyme-substrate complex bound together is an intermediate in a reaction, denoted by [ES].
\[ \text{Step 2}\;\;\;\;\; ES \rightarrow E + P \]
where P stands for products, E for enzyme, and S for substrate.
The rate determining step for an enzyme-substrate reaction is always the second step in which [ES] is converted into the product. The reason for this is because once the enzyme performs its duty, it is free to do more work. Once an enzyme can do more work after conversion, a reaction can go faster.
The rates of enzyme-substrate reactions oscillate between first order and second order. Initially, a reaction will be first order because it will depend on the amount of substrate added. When the maximum amount of active sites are consumed, the rate of an enzyme-substrate reaction maximizes, becoming a zero order reaction in which the rate of reaction is constant. A zero-order reaction is typically denoted graphically by an asymptote, which indicates the rate limit of the reaction. When an enzyme-substrate reaction tends toward zero order, the only way to make a reaction speed up is to add more enzyme, therefore adding more active sites. If more enzyme is added to a zero order maximized reaction, then the reaction will go back to first order until either all of the active sites are taken once again or all of the substrate is converted into product, leaving an excess of empty active sites.
The rate of production of the product depends on the velocity (V) of a reaction. For any enzyme-substrate reaction to go forward, the rate of product formation (or decomposition of ES) must equal the rate of formation of ES. If ES only forms and does not decompose into product, then the enzyme is not working. Enzymes that do not work are discussed later, and may be a result of faulty RNA translation from DNA, which causes the active site on an enzyme to be malformed.
Because the total concentration of an enzyme can never be accurately measured, biochemists like to use \(E_0\) to denote the sum of unbound enzyme versus bound enzyme ES. A convenient constant to use when relating the rates of the forward versus reverse reactions of enzyme chemistry is \(K_M\). This constant can be derived by dividing the rates of formation of unbound E (k-1 and k2) by the bound ES (k1).
\[ K_M=\dfrac{k_{-1}+k_2}{k_1} \]
where
- \(k_{-1}\) is \(ES \rightarrow E+S\)
- \(k_2\) is \(ES \rightarrow E+P\), and
- \(k_1\) is \(E+S \rightarrow ES\).
Note that this constant is always changing due to fluctuations in the rates of the forward and reverse reactions due to the concentrations of the enzyme or substrate. The equation for velocity can then be understood.
\[V=\dfrac{k_2[E_0][S]}{K_M+[S]}\]
If \([S]\) is low, then the value of \(K_M\) will be large, and the reaction rate will depend on the concentration of the substrate. This reaction will be a first order reaction because there is enough enzyme to drive the reaction forward at a relatively fast rate.
\[K_M>>[S]\]
If the concentration of the substrate is high and all of the active sites are taken, then \(K_M\) will be sufficiently less than [S], and the reaction will tend toward its maximum rate. More enzyme will need to be added to drive this reaction faster, and the reaction will become zero order and attain an asymptote.
\[K_M<<[S]\]
Thermodynamics
The thermodynamics of a biological reaction are crucial. Your body temperature stays at a constant 97.5 to 98.8 degrees Fahrenheit because a higher body temperature can cause certain proteins to denature in your body. A lowering of this range will cause reactions to slow down, which also may cause death. However, slight changes above this set homeostasis can drive all of the reactions forward in your body, causing you to burn more energy, which partially escapes in the form of heat. Therefore, when you have a fever, your mother or father may have felt your forehead to see if you were warmer than usual.
Active Site Chemistry
Active sites are the parts of enzymes that are substrate-specific. Certain enzymes will only bind to certain substrates because of a site resembling a lock-key on the surface of the enzyme. We will be taking a look at a very common enzyme family called serine protease as an example of how active site chemistry works. The serine protease family is an important enzyme for digestion, blood clotting, and fertilization. They are also the enzymes that catalyse peptide bond cleavage by attacking the carbonyl bond. Serine proteases are most famous for their specificity for substrates. They contain disulphide linkages (S--S) to keep their shape. Charged side chains are found on the outside of the enzyme, interacting with the solvent unless involved in catalysis.

Let us use an enzyme called trypsin in the serine protease family. Trypsin's active site has two domains, with the active site between the two. At the center of each domain is a barrel structure. Polar regions of the structure are well hydrated. Trypsin's active site contains the amino acid sequence Asp 102, His 57, Ser 195 (Aspartic Acid, Histidine, and Serine respectively). The numbers correspond with the actual sequence and position of the amino acids. These amino acids are found on loop regions of the two domains, and represent the charge relay system for the active site. The specificity pocket is also found in the loops of the two domains. These two domains of barrel structures are important because they provide a scaffold on which the specific amino acid bonds can interact to form the substrate-specific active site. The connection between the domains is less tight at the active site and may allow more rigid movements within the domains that may contribute to catalysis. These rigid body movements are a fundamental part of enzyme catalysis. We still have much to research on serine proteases because not much is understood about their crucial chemistry.

The Reaction of Trypsin
His 57 and Asp 102 are supposed to fix the Ser 195 to a state capable of reacting with the incoming peptide chain and to stabilize any intermediate formed during catalysis. His 57 acts as a strong base, abstracting the alcoholic proton of Ser 195 and moves it to the amine leaving group. The negative end of Asp 102 cancels out the positive charge developed by His 57 during the transition state. Then, the hydrolysis (adding of H2O) of the acyl-enzyme releases the product. (At this link, do not pay attention to the actual reaction, just pay attention to the highlighted intermediate of acyl-enzyme. This reaction is a protease inhibitor.)
The actual reaction mechanism. And another...
These catalysts drive the reaction forward 1,000,000 times faster than the reaction without a catalyst.
Problems:
1) What is a catalyst?
A catalyst is a compound that speeds up a reaction.
2) What is an enzyme?
An enzyme is a biological catalyst that speeds up reactions and interactions between molecules in biological systems.
3) What is the name of reactants that enter a substrate to form products at a faster rate?
The name of the reactants that enter a substrate to form products at a faster time are called substrates.
4) In which place on the enzyme does the substrate bind (to that enzyme, specifically) to give us products at a faster rate?
The name of the place on which the substrate binds is the active site of the enzyme.
Outside Links
- http://www.microtack.com/html/enzyme1.htm - How Do Enzymes Function?
- www.buzzle.com/articles/enzym...e-complex.html - More about enzyme-substrates
- http://www.tutorvista.com/content/ch...-catalysis.php
- http://highered.mcgraw-hill.com/sites/0072495855/student_view0/chapter2/animation__how_enzymes_work.html -This site has a brief clip about enzymes, if you are a better listener than reader. It also has a short quiz with answers on the page, as well.
Reference
- Suckling, Gibson, and Andrew Pitt. Enzyme Chemistry: Impact and Applications. Third Edition. Blackie Academic and Professional: Glasgow, UK. 1998. pp. 92-95.
- Keleti, T. Basic Enzyme Kinetics. Akademiai Kiado: Budapest, Hungary. 1986. pp. 19-30.
- http://www.rcsb.org
Contributors and Attributions
- Michael Abdelnour (UCD), Amber Quave (UCD), Thomas Ma (UCD), Hygan Baghoyan (UCD)

