Horseradish Peroxidase
- Page ID
- 400
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Horseradish Peroxidase is a metalloenzyme that exists in the root of the horseradish plant. There are a large number of peroxidase isoenzymes of horseradish with the most common being the C type.1 This type will be discussed in the remaining report because of the extensive research that has been completed on it.
Introduction
Horseradish peroxidase uses hydrogen peroxide to oxidize both organic and inorganic compounds.1 Horseradish peroxidase along with other heme peroxidases are brightly colored especially under the near-ultraviolet light.2 This property of heme peroxidases make them useful for attaching to “transparent” proteins so that they can be seen under different wavelengths. The heme group that is in horseradish peroxidase is simpler than those in mammalians and therefore makes it an excellent starting point in the in-depth study of heme peroxidases and their functions.2 It has been found that horseradish peroxidase when combined with other compounds is highly reactive toward human tumor cells. A better understanding of horseradish peroxidase could lead to a new targeted cancer therapy.1
Structure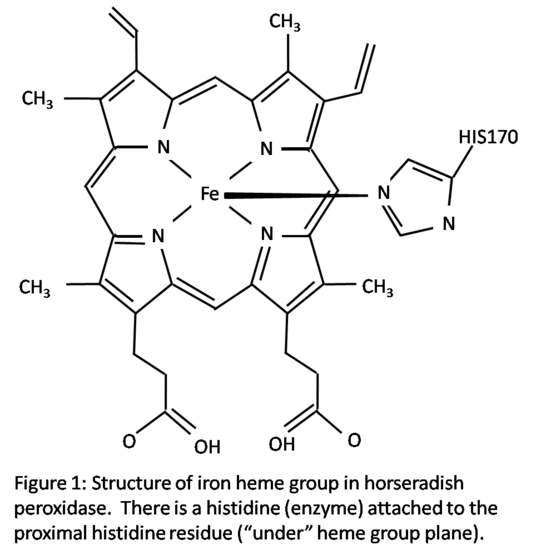
Horseradish peroxidase C has two metal centers, one of iron heme group and two calcium atoms.1 The structure is shown in Figure 1. The heme group has a planar structure with the iron atom held tightly in the middle of a porphyrin ring which is comprised of four pyrrole molecules.3 Iron has two open bonding sites, one above and one below the plane of the heme group. The heme group has a histidine (enzyme) attached in the proximal histidine residue (His170) which is located below the heme group in Figure 1. The second histidine residue in the distal side of the heme group, above the heme group, is vacant in the resting state.1 This site is open for hydrogen peroxide to attach during reduction-oxidation reactions. An oxygen atom will bond to this vacant site during activation. The iron atom’s sixth octahedral position is considered the active site of the enzyme. During the enzyme reaction, the bonding of the hydrogen peroxide to the iron atom creates an octahedral configuration around the iron atom. Other small molecules can also bond to the distal site, creating the same octahedral configuration.1
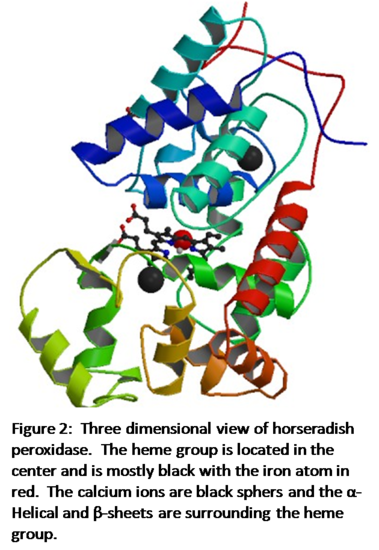
Figure 2 shows the three-dimensional structure of horseradish peroxidase.4 The iron heme is in the center of the enzyme shown in black with the iron atom as the red sphere. The two calcium atoms are black spheres and lie within the helical regions of the enzyme, with one in the distal region and one in the proximal region. The α-Helical and β-sheet regions of the enzyme are shown as the multicolored helical structures. According to Veitch, “Each calcium site is seven-coordinate with oxygen-donor ligands provided by a combination of amino acid side-chain carboxylates (Asp), hydroxyl groups (Ser, Thr), backbone carbonyls and a structural water molecule (distal site only).”1 Both the heme group and the calcium atoms are crucial to the enzyme working properly and the loss of one would result in instability.
Symmetry and Raman Spectrometry
Pure iron, at room temperature, has a body-centered cubic structure. However, according to the RCSB Protein Data Bank, x-ray diffraction shows that horseradish peroxidase has an orthorhombic structure.4 An orthorhombic structure is where all of the angles in the unit cell equal 90o and each of the three sides do no equal each other. In the case of horseradish peroxidase the three side lengths are: a=40.28 Å, b=67.46 Å, and c=117.11 Å.4 X-ray diffraction also provided the data to create the three-dimensional structure in Figure 2.
The immediate ligand environment of the iron atom in the resting state boasts a point group of D4d. The molecule is not linear nor does it have tetrahedral, icosahedral, or octahedral symmetry (at least in the rest state). The molecule has a C4 rotation axis and has 4 C2 axes perpendicular to the C4 axis.There is not a σh plane due to the fifth nitrogen atom which is part of the His170 residue that is bonded to the bottom of the iron atom.There are 4 σv planes that contain the C4 axis which leads to a point group symmetry of D4d. When the metalloenzyme is not in the resting state the sixth octahedral position of the iron atom is filled which will lead to a change in point group designation.Figure 2 shows the structure of the entire metalloenzyme.It can be seen that there is no symmetry within this structure and horseradish peroxidase as a whole has a point group of C1. There is no principal axis, mirror plane, or center of inversion in the enzyme.
Resonance Raman techniques are difficult to use with horseradish peroxidase due to the reactivity and photolability of the enzyme. This causes the horseradish peroxidase enzyme to break down into a second phase (HRP-II), ferric, and ferrous species.5 Scientists W. Anthony Oertling and Gerald T. Babcock attempted to mitigate this problem by lowering to cryogenic temperatures, pulsing a near-UV laser, and mixing hydrogen peroxide in the enzyme very quickly. Although some of the old problems still arose, they were able to get a Raman spectrum of the horseradish peroxidase. Horseradish peroxidase has three intermediate compounds all of which have a different color under optical spectroscopy.2 Because of this, the different stages of the redox cycle can be identified and a better understand of what constitutes each part of the cycle can be found.
Reactions
Figure 3 shows the five oxidation states of horseradish peroxidase.6 During the redox reaction, the hydrogen peroxide bonds to the vacant octahedral position on the iron atom which initiates the reaction. There are three different intermediate horseradish peroxidase compounds that form during the reaction. They are created as shown in the figure, with either an addition of an electron or a reaction with hydrogen peroxide. The reduction of compound I to compound II and compound II back to the rest state is carried out by reduction substrates. They are usually phenols or aromatic amines.2 To fully understand how the structure is changing during these stages, x-ray diffraction data must be taken. This, however, has posed a problem because electrons that are stirred up by the x-rays will alter the reduction-oxidation stage of the active site.6
References
- Veitch, Nigel C., Horseradish peroxidase: a modern view of a classic enzyme, Phytochemistry, 65, 2004, 249-259, 23-May-2010
- Dunford, H.Brian and Jones, Peter A., Peroxidases and Catalases: Biochemistry, Biophysics, Biotechnology and Physiology, 2nd Edition, 2010, John Wiley and Sons Inc.
- “Hemoglobin” School of Chemistry- Bristol University-UK, Web. 23-May-2010, <http://www.chm.bris.ac.uk/motm/hemoglobin/hemoglobjm.htm>
- “Structure of Horseradish Peroxidase C1A Compound I”, RCSB Protein Data Bank, 2010, 23-May-2010, <http://www.pdb.org/pdb/explore/explore.do?structureId=1HCH>
- Oertling, W. Anthony, Resonance Raman Scattering from Horseradish Peroxidase Compound I, Journal of the American Chemical Society, 1985, 107 (22), 6406-6407, 23-May-2010
- Berglund, Gunnar I., Carlsson, Gunilla H., et al., The catalytic pathway of horseradish peroxidase at high resolution, Nature, 2002, 417, 463-468, 23-May-2010


