3. Proteins as Enzymes
- Page ID
- 3964
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)This page is an introduction to how proteins can work as enzymes - biological catalysts.
Enzymes as catalysts
Enzymes are mainly globular proteins - protein molecules where the tertiary structure has given the molecule a generally rounded, ball shape (although perhaps a very squashed ball in some cases). The other type of proteins (fibrous proteins) have long thin structures and are found in tissues like muscle and hair. We aren't interested in those in this topic.
These globular proteins can be amazingly active catalysts. You are probably familiar with the use of catalysts like manganese(IV) oxide in decomposing hydrogen peroxide to give oxygen and water. The enzyme catalase will also do this - but at a spectacular rate compared with inorganic catalysts. One molecule of catalase can decompose almost a hundred thousand molecules of hydrogen peroxide every second. That's very impressive! This is a model of catalase, showing the globular structure - a bit like a tangled mass of string:
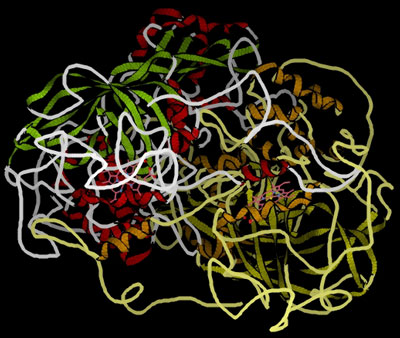
An important point about enzymes is that they are very specific about what they can catalyse. Even small changes in the reactant molecule can stop the enzyme from catalysing its reaction. The reason for this lies in the active site present in the enzyme . . .
Active sites
Active sites are cracks or hollows on the surface of the enzyme caused by the way the protein folds itself up into its tertiary structure. Molecules of just the right shape, and with just the right arrangement of attractive groups (see later) can fit into these active sites. Other molecules won't fit or won't have the right groups to bind to the surface of the active site.
The usual analogy for this is a key fitting into a lock. For the key to work properly it has to fit exactly into the lock.
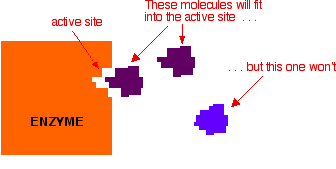
In chemistry, we would describe the molecule which is actually going to react (the purple one in the diagram) as the reactant. In biology and biochemistry, the reactant in an enzyme reaction is known instead as the substrate.
You mustn't take this picture of the way a substrate fits into its enzyme too literally. What is just as important as the physical shape of the substrate are the bonds which it can form with the enzyme.
Enzymes are protein molecules - long chains of amino acid residues. Remember that sticking out all along those chains are the side groups of the amino acids - the "R" groups that we talked about on the page about protein structure.
Active sites, of course, have these "R" groups lining them as well - typically from about 3 to 12 in an active site. The next diagram shows an imaginary active site:

Remember that these "R" groups contain the sort of features which are responsible for the tertiary structure in proteins. For example, they may contain ionic groups like -NH3+ or -COO-, or -OH groups which can hydrogen bond, or hydrocarbon chains or rings which can contribute to van der Waals forces.
Groups like these help a substrate to attach to the active site - but only if the substrate molecule has an arrangement of groups in the right places to interact with those on the enzyme.
The diagram shows a possible set of interactions involving two ionic bonds and a hydrogen bond.
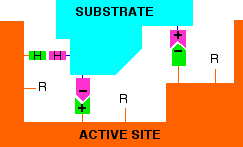
The groups shown with + or - signs are obvious. The ones with the "H"s in them are groups capable of hydrogen bonding. It is possible that one or more of the unused "R" groups in the active site could also be helping with van der Waals attractions between them and the substrate.
If the arrangement of the groups on the active site or the substrate was even slightly different, the bonding almost certainly wouldn't be as good - and in that sense, a different substrate wouldn't fit the active site on the enzyme.
This process of the catalyst reacting with the substrate and eventually forming products is often summarised as:
![]()
. . . where E is the enzyme, S the substrate and P the products.
The formation of the complex is reversible - the substrate could obviously just break away again before it converted into products. The second stage is shown as one-way, but might be reversible in some cases. It would depend on the energetics of the reaction.
So why does attaching itself to an enzyme increase the rate at which the substrate converts into products?
It isn't at all obvious why that should be - and most sources providing information at this introductory level just gloss over it or talk about it in vague general terms (which is what I am going to be forced to do, because I can't find a simple example to talk about!).
Catalysts in general (and enzymes are no exception) work by providing the reaction with a route with a lower activation energy. Attaching the substrate to the active site must allow electron movements which end up in bonds breaking much more easily than if the enzyme wasn't there.
Strangely, it is much easier to see what might be happening in other cases where the situation is a bit more complicated . . .
Enzyme cofactors
What we have said so far is a major over-simplification for most enzymes. Most enzymes aren't in fact just pure protein molecules. Other non-protein bits and pieces are needed to make them work. These are known as cofactors.
In the absence of the right cofactor, the enzyme doesn't work. For those of you who like collecting obscure words, the inactive protein molecule is known as an apoenzyme. When the cofactor is in place so that it becomes an active enzyme, it is called a holoenzyme.
There are two basically different sorts of cofactors. Some are bound tightly to the protein molecule so that they become a part of the enzyme - these are called prosthetic groups.
Some are entirely free of the enzyme and attach themselves to the active site alongside the substrate - these are called coenzymes.
Prosthetic groups
Prosthetic groups can be as simple as a single metal ion bound into the enzyme's structure, or may be a more complicated organic molecule (which might also contain a metal ion). The enzymes carbonic anhydrase and catalase are simple examples of the two types.
The ideal gas law is easy to remember and apply in solving problems, as long as you get the proper values a
Zinc ions in carbonic anhydrase
Carbonic anhydrase is an enzyme which catalyses the conversion of carbon dioxide into hydrogencarbonate ions (or the reverse) in the cell. (If you look this up elsewhere, you will find that biochemists tend to persist in calling hydrogencarbonate by its old name, bicarbonate!)
![]()
In fact, there are a whole family of carbonic anhydrases all based around different proteins, but all of them have a zinc ion bound up in the active site. In this case, the mechanism is well understood and simple. We'll look at this in some detail, because it is a good illustration of how enzymes work.
The zinc ion is bound to the protein chain via three links to separate histidine residues in the chain - shown in pink in the picture of one version of carbonic anhydrase. The zinc is also attached to an -OH group - shown in the picture using red for the oxygen and white for the hydrogen.

The structure of the amino acid histidine is . . .

. . . and when it is a part of a protein chain, it is joined up like this:

If you look at the model of the arrangement around the zinc ion in the picture above, you should at least be able to pick out the ring part of the three molecules. The zinc ion is bound to these histidine rings via dative covalent (co-ordinate covalent) bonds from lone pairs on the nitrogen atoms. Simplifying the structure around the zinc:
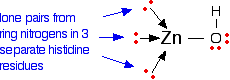
The arrangement of the four groups around the zinc is approximately tetrahedral. Notice that I have distorted the usual roughly tetrahedral arrangement of electron pairs around the oxygen - that's just to keep the diagram as clear as possible.
So that's the structure around the zinc. How does this catalyse the reaction between carbon dioxide and water?
A carbon dioxide molecule is held by a nearby part of the active site so that one of the lone pairs on the oxygen is pointing straight at the carbon atom in the middle of the carbon dioxide molecule. Attaching it to the enzyme also increases the existing polarity of the carbon-oxygen bonds.

If you have done any work on organic reaction mechanisms at all, then it is pretty obvious what is going to happen. The lone pair forms a bond with the carbon atom and part of one of the carbon-oxygen bonds breaks and leaves the oxygen atom with a negative charge on it.

What you now have is a hydrogencarbonate ion attached to the zinc.
The next diagram shows this broken away and replaced with a water molecule from the cell solution.

All that now needs to happen to get the catalyst back to where it started is for the water to lose a hydrogen ion. This is transferred by another water molecule to a nearby amino acid residue with a nitrogen in the "R" group - and eventually, by a series of similar transfers, out of the active site completely. . . . and the carbonic anhydrase enzyme can do this sequence of reactions about a million times a second. This is a wonderful piece of molecular machinery!
The heme (US: heme) group in catalase
Remember the model of catalase from further up the page . . .
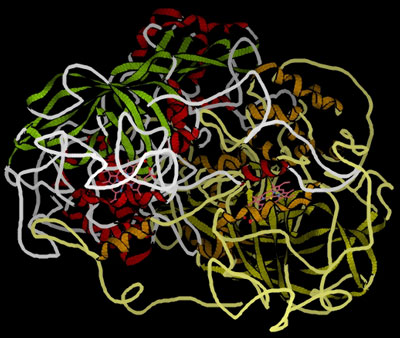
At the time, I mentioned the non-protein groups which this contains, shown in pink in the picture. These are heme (US: heme) groups bound to the protein molecule, and an essential part of the working of the catalase. The heme group is a good example of a prosthetic group. If it wasn't there, the protein molecule wouldn't work as a catalyst. The heme groups contain an iron(III) ion bound into a ring molecule - one of a number of related molecules called porphyrins. The iron is locked into the centre of the porphyrin molecule via dative covalent bonds from four nitrogen atoms in the ring structure. There are various types of porphyrin, so there are various different heme groups. The one we are interested in is called heme B, and a model of the heme B group (with the iron(III) ion in grey at the centre) looks like this:

The reaction that catalase carries out is the decomposition of hydrogen peroxide into water and oxygen.
![]()
A lot of work has been done on the mechanism for this reaction, but I am only going to give you a simplified version rather than describe it in full. Although it looks fairly simple on the surface, there are a lot of hidden things going on to complicate it.
Essentially the reaction happens in two stages and involves the iron changing its oxidation state. An easy change of oxidation state is one of the main characteristics of transition metals. In the lab, iron commonly has two oxidation states (as well as zero in the metal itself), +2 and +3, and changes readily from one to the other.
In catalase, the change is from +3 to the far less common +4 and back again.
In the first stage there is a reaction between a hydrogen peroxide molecule and the active site to give:
![]()
The "Enzyme" in the equation refers to everything (heme group and protein) apart from the iron ion. The "(III)" and "(IV)" are the oxidation states of the iron in both cases. This equation (and the next one) are NOT proper chemical equations. They are just summaries of the most obvious things which have happened.
The new arrangement around the iron then reacts with a second hydrogen peroxide to regenerate the original structure and produce oxygen and a second molecule of water.
![]()
What is hidden away in this simplification are the other things that are happening at the same time - for example, the rest of the heme group and some of the amino acid residues around the active site are also changed during each stage of the reaction.
And if you think about what has to happen to the hydrogen peroxide molecule in both reactions, it has to be more complicated than this suggests. Hydrogen peroxide is joined up as H-O-O-H, and yet both hydrogens end up attached to the same oxygen. That is quite a complicated thing to arrange in small steps in a mechanism, and involves hydrogen ions being transferred via amino acids residues in the active site.
So do you need to remember all this for chemistry purposes at this level? No - not unless your syllabus specifically asks you for it. It is basically just an illustration of the term "prosthetic group".
It also shows that even in a biochemical situation, transition metals behave in the same sort of way as they do in inorganic chemistry - they form complexes, and they change their oxidation state. And if you want to follow this up to look in detail at what is happening, you will find the same sort of interactions around the active site that we looked at in the simpler case of carbonic anydrase. (But please don't waste time on this unless you have to - it is seriously complicated!)
Coenzymes
Coenzymes are another form of cofactor. They are different from prosthetic groups in that they aren't permanently attached to the protein molecule. Instead, coenzymes attach themselves to the active site alongside the substrate, and the reaction involves both of them. Once they have reacted, they both leave the active site - both changed in some way. A simple diagram showing a substrate and coenzyme together in the active site might look like this:

It is much easier to understand this with a (relatively) simple example.
NAD+ as coenzyme with alcohol dehydrogenase
Alcohol dehydrogenase is an enzyme which starts the process by which alcohol (ethanol) in the blood is oxidised to harmless products. The name "dehydrogenase" suggests that it is oxidising the ethanol by removing hydrogens from it.
The reaction is actually between ethanol and the coenzyme NAD+ attached side-by-side to the active site of the protein molecule. NAD+ is a commonly used coenzyme in all sorts of redox reactions in the cell.
NAD+ stands for nicotinamide adenine dinucleotide. The plus sign which is a part of its name is because it carries a positive charge on a nitrogen atom in the structure.
The "nicotinamide" part of the structure comes from the vitamin variously called vitamin B3, niacin or nicotinic acid. Several important coenzymes are derived from vitamins.
Ethanol is oxidised by a reaction with NAD+ helped by the active site of the enzyme. At the end of the reaction, ethanal (acetaldehyde) is formed, and the NAD+ has been converted into another compound known as NADH.

As far as the NAD+ is concerned, it has picked up a hydrogen atom together with an extra electron which has neutralised the charge. Both major products - ethanal and NADH - leave the active site and are processed further in other cell reactions.
The very poisonous ethanal is oxidised at once to ethanoic acid using a different enzyme, but again using NAD+ as the coenzyme. And the ethanoic acid from that reacts on through a whole set of further enzyme-controlled reactions to eventually end up as carbon dioxide and water.
What about the NADH? This is a coenzyme in its own right, and takes part in reactions where something needs reducing. The hydrogen atom and the extra electron that it picked up from the ethanol are given to something else. In the process, of course, the NADH gets oxidised back to NAD+ again. In general terms, for a substrate S which needs reducing:
![]()


