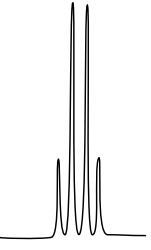
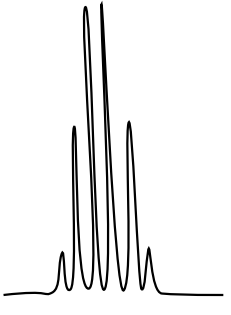
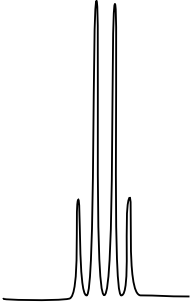
10: ¹H NMR
- Page ID
- 332810
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)1H - NMR Spectroscopy Analysis
Overview
Four types of information are found in 1H NMR spectra:
- Number of different types of protons (symmetry)
- Chemical Shift (bonding environment)
- Integration (ratio of protons per resonance)
- Multiplicity [n+1] (number of neighboring protons)
- How is this similar to 13C NMR? DEPT?
- How is this different from 13C NMR?
1H NMR Shift and Symmetry
1. Symmetry
Like 13C, only the unique H atoms will provide a different signal in the spectrum. A peak for every unique H atom will show up.
- Look for symmetry and “hidden” H atoms.
- For example:
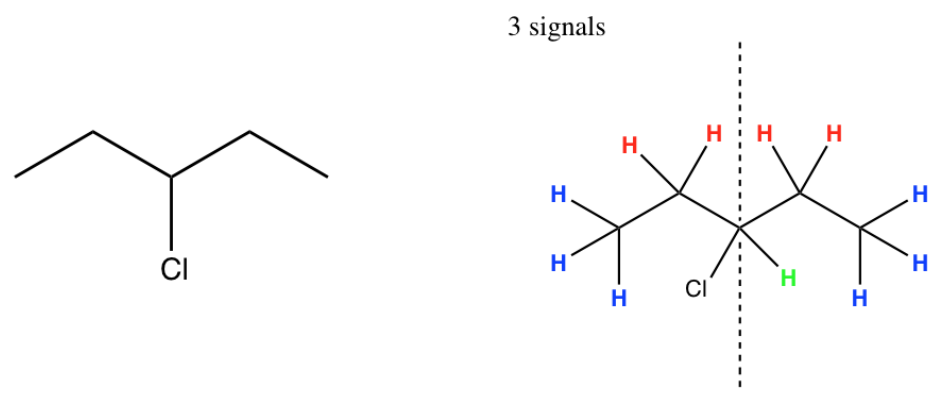
- Predict the number of peaks that will show up in the spectra of the following compounds:

1H NMR Chemical Shift
2. Chemical Shift (bonding environment)
Like 13C, Chemical Shift (\(\delta\)) is the x-axis.
– Units = ppm
– Scale is 0 – 14 ppm compared to ___________ ppm for 13C
– Depends on Geometry & Dipole
Chemical Shift and Geometry
sp3 is tetrahedral (carbon with 4 single bonds)
sp2 is trigonal planar (carbon with two single bonds and one double bond)
In proton NMR spectroscopy, the shift of the proton is affected by the geometry of the carbon to which it is bonded.
- Predict where a proton bonded to an sp2 carbon would show up.
- Predict where a proton bonded to an sp3 carbon would show up.

1H NMR Shift and Symmetry
Chemical Shift and Dipole
As the proton of a functional group becomes more partially positive, it will appear further downfield (larger ppm).
- Order the following functional groups by the dipole strength. (1 = proton that feels the most neutral; 5 = proton that feels the most positive.)

1H Shifts of Alkanes*
* All spectra are either from SDBS (Japan National Institute of Advanced Industrial Science and Technology) or simulated.
Hydrocarbons
- All of the hydrogens are bonded to [ sp3 or sp2 ] carbons.
- Based on the hybridization where would you expect the peaks to appear?
Here is a spectrum of a hydrocarbon.

- Without the presences of any dipoles in the structure, the hydrogens appear below _______ ppm.
1H Shifts of Alkyls next to Carbonyls and Electronegative Atoms
Hydrogens bonded to sp3 carbons with electron-withdrawing functional groups are shifted downfield (to the left).
2-butanone
- The hydrogens on the carbons next to the carbonyl group are shifted to _______ppm.

1-chloro-propane
- The hydrogens on the carbons next to an electronegative atom (O, N, Cl, Br) are shifted to _______ ppm.
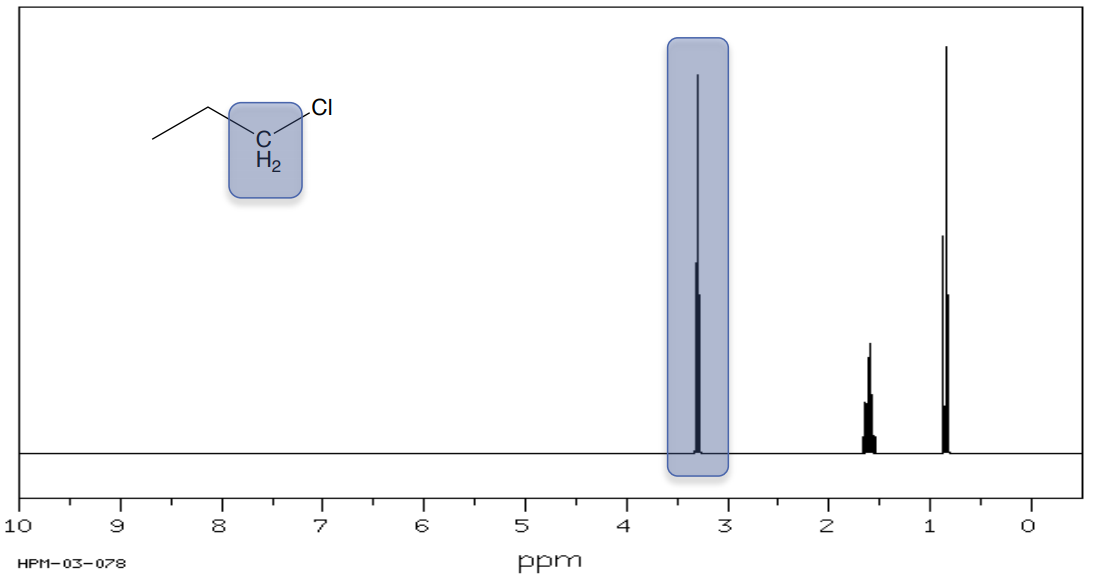
1H Shifts of Alkyls next to Carbonyls and Electronegative Atoms
- Based on the shifts, assign the methyl groups to the correct peak in the proton NMR spectrum.


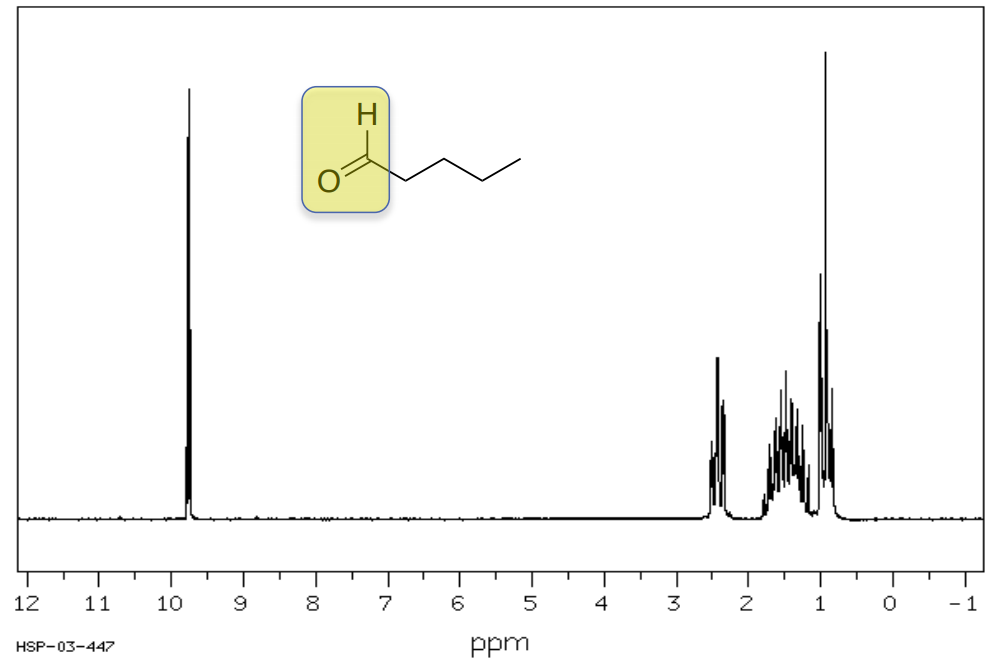
1H Shifts of Aldehydes and Acids
Pentanal
Typically, hydrogens on aldehydes appear between 8-10 ppm.
- Circle or highlight the peaks corresponding to the highlighted atoms.
Benzoic Acid
Typically, hydrogens on carboxylic acids appear between 11-14 ppm.
- Circle or highlight the peaks corresponding to the highlighted atoms.
- Bonus: Identify the peaks associated with hydrogens on the aromatic ring.

1H Shifts of Alkenes and Aromatics
2-hexene
- Typically, hydrogens on an alkene appear between 4-6 ppm.
- Carbons involved in a double bond are [ sp3 or sp2 ] carbons.
- Circle or highlight the peaks corresponding to the highlighted atoms

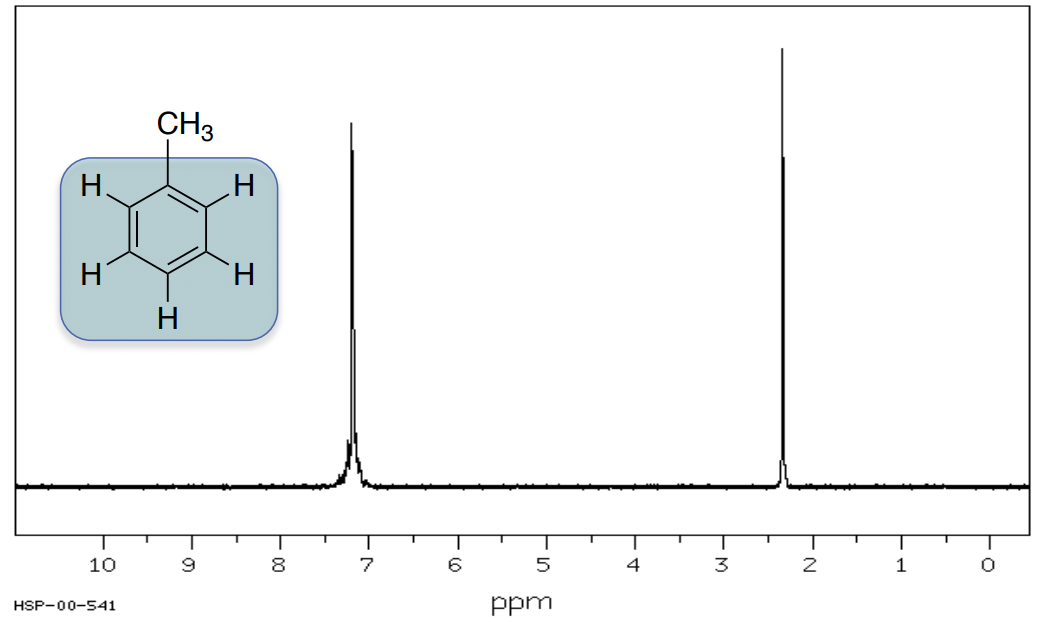
Methylbenzene
Typically, hydrogens on an aromatic ring appear between 6-8 ppm.
- Circle or highlight the peaks corresponding to the highlighted atoms.
1H NMR shift: Anisotropy
Review shifts:
- Electron-donating groups provide a shielding effect which moves the shift of proton [ upfield / downfield ].
- Proton shifts move [ upfield / downfield ] when electronegative substituents are attached to the same or an adjacent carbon causing the proton to be deshielded.
The local circulation of electrons creates a magnetic field which provides a shielding effect, magnetic anisotropy.
Anisotropy effects are usually used to explain shifts of sp2 and sp hybridized systems.
Protons on alkenes and aromatic rings behave as if they were bonded to electronegative atoms (~5-8 ppm).

- The local circulation of electrons around an aromatic ring creates a magnetic field aligned with the external field deshielding the protons which causes the shift to move [ upfield / downfield ].
In contrast, protons on alkynes are not shifted very far (~2-3 ppm).

- Proton shifts move [ upfield / downfield ] when the magnetic field is aligned opposite to the external field providing a shielding effect.
1H NMR Shift: Solvent Effects
The chemical shift of a given proton will change depending on the environment. The variability is especially large for NH and OH protons (several ppm).
Different solvents can cause changes in chemical shifts as large as 0.5 to 0.8 ppm

1-phenylbutan-1-one in deuterated benzene vs deuterated chloroform
- Suggest a reason that solvent can impact chemical shift. Consider how it might make an atom ‘feel’ more or less positive.
1H NMR Shift: Hydrogen Bonding & Concentration
Chemical shifts of protons can vary with concentration, especially if hydrogen bonding can occur. The chemical shifts of protons on oxygen (OH) and nitrogen (NH), which are often directly involved in hydrogen bonding are especially strongly dependent (several ppm) on concentration, solvent and temperature.

- Explain why hydrogen bonding would affect chemical shift. In other words, would a proton that is involved in hydrogen bonding, feel more or less deshielded?
- Explain why concentration will affect the amount of hydrogen bonding.
Summary of Chemical Shifts in 13C NMR vs 1H NMR
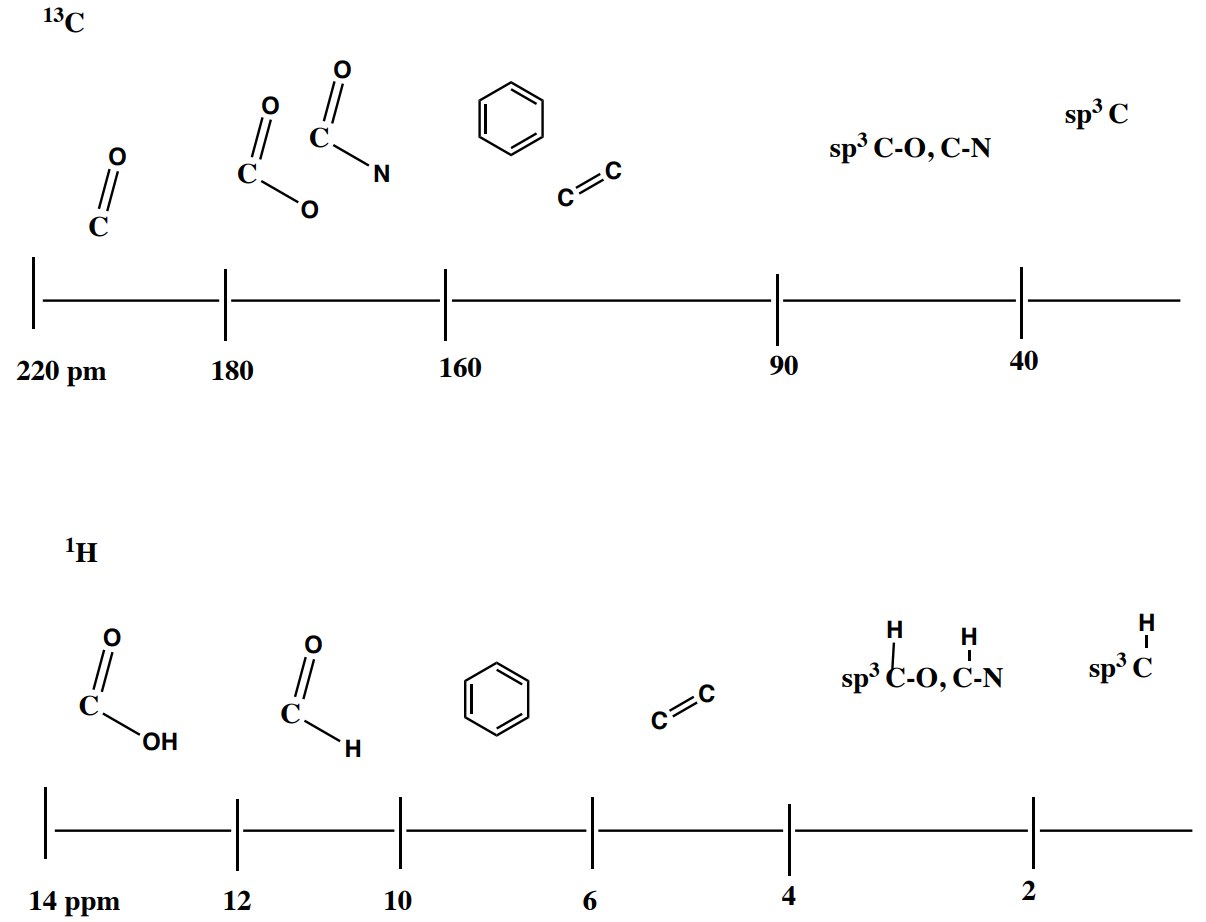
1H NMR Integration
The area under an NMR peak is proportional to the number of hydrogens which that resonance represents.
Measuring or integrating the different NMR resonances provides information regarding the relative numbers of different hydrogens, not the absolute number.
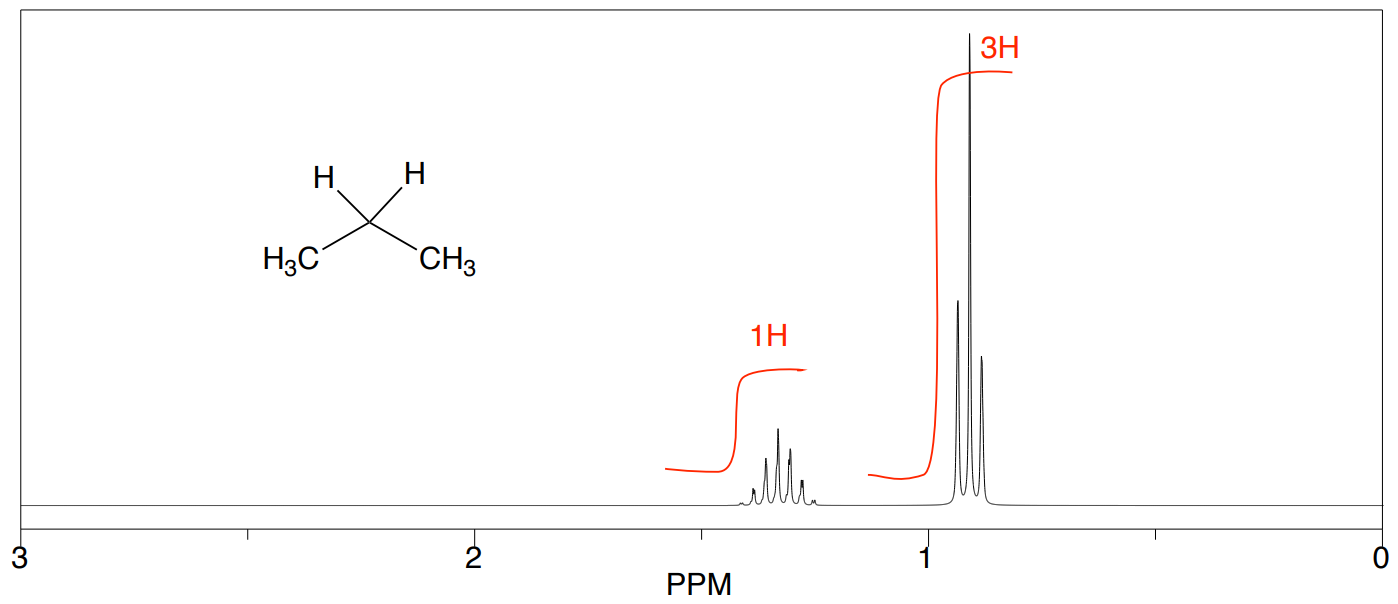
Propane
- How many unique hydrogens in propane?
- What is the ratio of hydrogens?
Experimentally, the integrals will usually appear as a line over the NMR spectrum. The heights of the “stair steps” can be directly compared as ratios to determine the ratio of hydrogen atoms represented by this peak.
- Does your prediction match the spectrum below?
1H NMR Integration Practice
- Which of the following would be possible hydrogen ratios represented by the integration curve shown below? Circle all that apply.
- 25:10:25
- 4:10:4
- 2:1:2
- 5:2:5

- Which of the following compounds match the integration ratio and chemical shift shown in this spectrum?



- CH3CH2OCH2CH3
- CH3CH2OH
1H NMR Multiplicity
4. Multiplicity (also called coupling or splitting)
Multiplicity usually follows a simple n+1 rule for the number of lines displayed for a peak.
| Pattern | Multiplicity | Number of H on C next door | Example |
|---|---|---|---|
 |
singlet (s) | 0 |  |
|
1:1 |
doublet (d) | 1 |  |
|
1:2:1 |
triplet (t) | 2 |  |
|
1:3:3:1 |
quartet (q) | 3 |  |
- How many lines would you expect to see for a proton when there are 6 H on the neighboring carbons?
- How many lines would you expect to see for a proton when there are 42 H on the neighboring carbons?
1H NMR Multiplicity Practice
Instead of predicting the number of lines, we often need to look at the peak and predict how many H are on neighboring carbons.
- Determine how many H are next door to the observed H looking at the following splitting patterns:
1H NMR Multiplicity
Here is the spectrum of propane again.
- Explain the multiplicity of each peak.

Here is the spectrum of ethanol. 
- Assign each peak in the spectrum to the correct set of hydrogens.
- Explain the multiplicity of each peak.

1H Spectral Analysis
- Predict the 13C NMR spectra for 2-butanone.
- Estimate appropriate shifts.

- Predict the 1H NMR spectra for 2-butanone.
- Estimate appropriate shifts.
- Include the correct multiplicity for each peak.
- Draw in the expected integration lines.

1H Spectral Analysis: Data Tables
Making data tables is important in structure elucidation. It helps with interpretation, keeping track of details, and communicating your analysis process (to your instructor or in a publication).
There is a standard format for 1H NMR tables. This format shows all four pieces of data that can be inferred from a spectrum (# peaks, shift, integration, multiplicity). Then in the final column, there is a place for you to show a partial structure derived from the data presented.
In the partial structure, the hydrogens being observed are underlined.

Partial Structures
When analyzing proton NMR spectra (and later 2D spectra), we will refer to part structures/spin systems.
For example, consider 3-heptanone:

Protons a, b, c and d constitute one spin system, an unbroken network of coupled protons. The ethyl group, e and f, constitutes a second, separate spin system, because there is no coupling between a and e, across the carbonyl.

- Complete the following table:
| Shift (ppm) | Integration | Multiplicity | Designation | Partial Structure |
|---|---|---|---|---|
| .9 | 3H | t | d | CH3-CH2 |
| 1.1 | t | f | ||
| 1.4 | 2H | |||
| 1.6 | ||||
| 2.4 | 2H | t | e | CO-CH2-CH3 |
| 2.45 |
1H Spectral Analysis: Data Tables
- Complete the following table to interpret this spectrum.
- Propose a structure for the compound.
C3H8O SU: ___________

| Shift (ppm) | Integration | Multiplicity | Partial Structure |
|---|---|---|---|
| 4.0 | 1H | sept | |
| 2.1 | 1H | s | |
| 1.2 | 6H | d |
| Proposed Structure: |
1H Spectral Analysis: Data Tables
- Complete the following table to interpret this spectrum.
- Propose a structure for the compound.
C3H8O SU: ___________
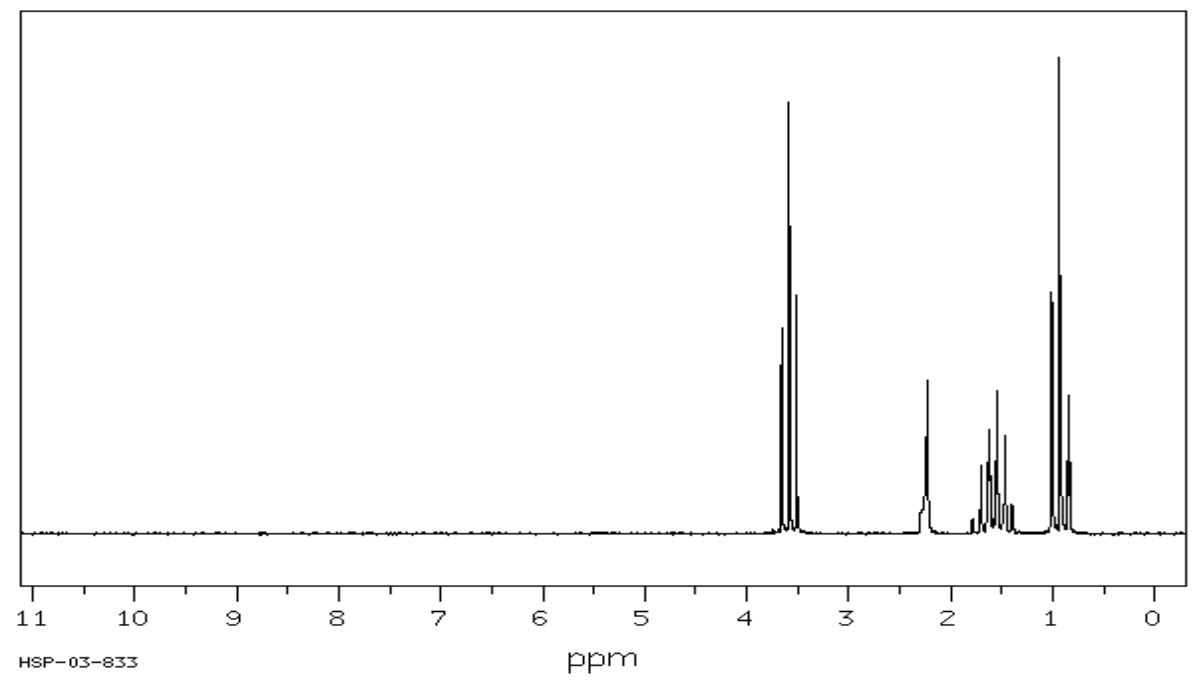
| Shift (ppm) | Integration | Multiplicity | Partial Structure |
|---|---|---|---|
| 3.6 | |||
| 2.4 | |||
| 1.5 | |||
| 1 |
| Proposed Structure: |
1H Spectral Analysis: Data Tables
- Complete the following table to interpret this spectrum.
- Propose a structure for the compound.
C3H7Br SU: ___________

| Shift (ppm) | Integration | Multiplicity | Partial Structure |
|---|---|---|---|
| 1.6 | |||
| 4.3 |
| Proposed Structure: |
1H Spectral Analysis: Data Tables
- Complete the following table to interpret this spectrum.
- Propose a structure for the compound.
C4H8O SU: ___________
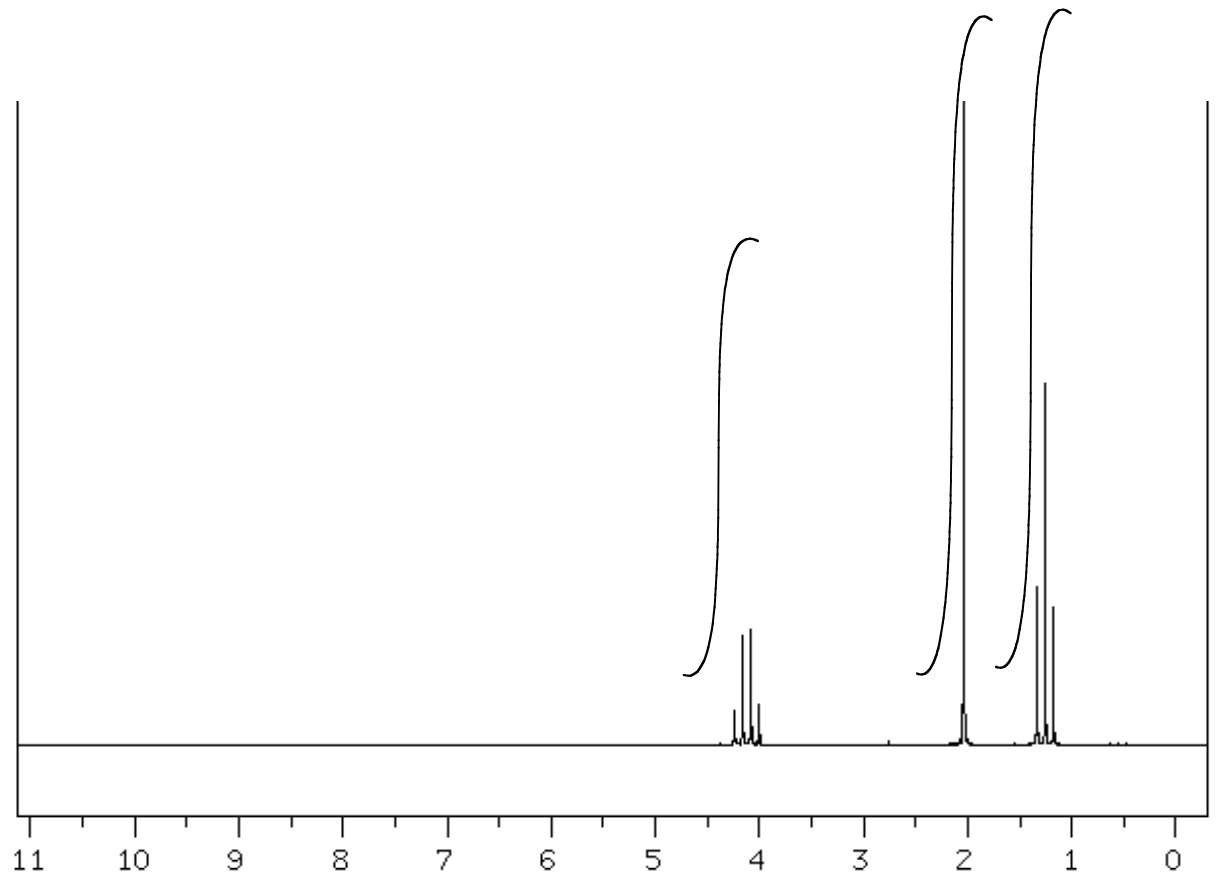
| Shift (ppm) | Integration | Multiplicity | Partial Structure |
|---|---|---|---|
| 4.05 | |||
| 2.0 | |||
| 1.2 |
| Proposed Structure: |
Aromatic Splitting Patterns
There are three isomers with the condensed formula CH3OC6H4CO2H. The C6H4 refers to a benzene, a ring of carbons with three double bonds.
- Draw the three isomers and predict what the 1H NMR spectrum will look like by filling in a data table.
Ortho Isomer structure:
| shift | integration | multiplicity | assignment |
Meta isomer structure:
| shift | integration | multiplicity | assignment |
Para isomer structure:
| shift | integration | multiplicity | assignment |
- The calculated spectra of these three isomers are on this page. Match the structure with the compound.



1H Spectral Problems
A. C5H10O2 SU: _____________

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
B. C5H10O2 SU: _____________

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
C. C9H10O SU: _____________

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
D. C10H12O SU: _____________

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
E. C5H10O SU: _____________

| Chemical Shift | Type of Bonding Environment |

| Proposed Structure: |
F. C5H10O2 SU: _____________
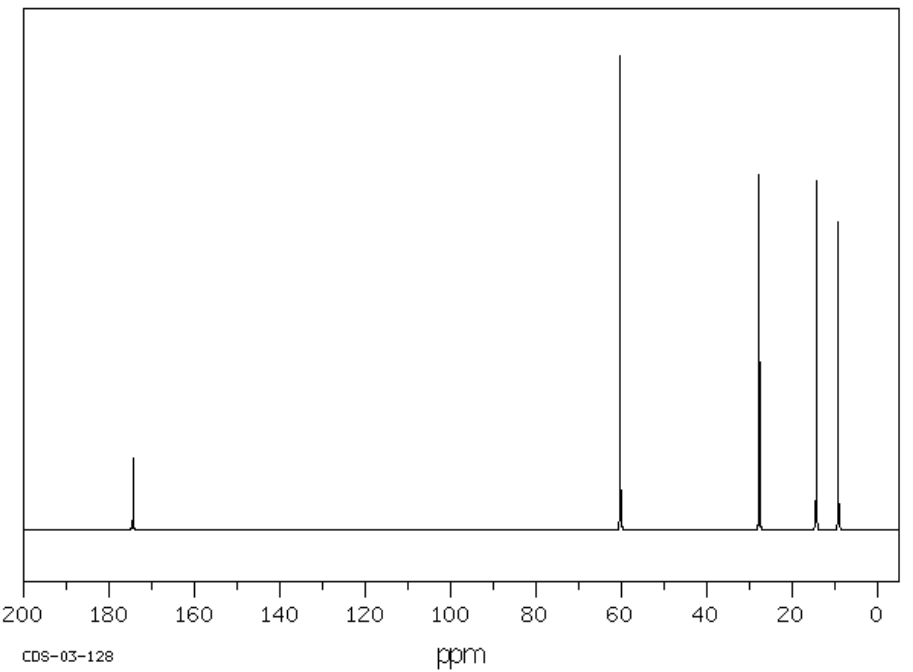
| Chemical Shift | Type of Bonding Environment |

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
G. C5H10O2 SU: _____________

| Chemical Shift | Type of Bonding Environment |

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
H. C5H10O SU: _____________
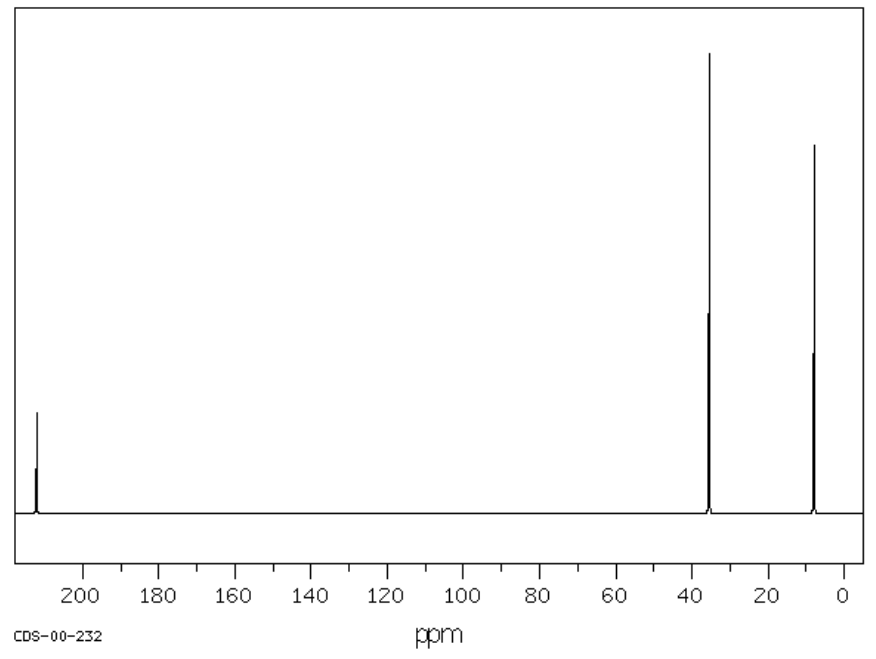
| Chemical Shift | Type of Bonding Environment |

| Chemical Shift | Integration | Multiplicity | Interpretation |
| Proposed Structure: |
I. C5H10O SU: _____________

| Chemical Shift | Type of Bonding Environment |

| Proposed Structure: |