Cross-Linking in DNA
- Page ID
- 50838
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)DNA is one of the most productive biomolecules in terms of purpose and replication. Its genes send out instructions to RNA, amino acids, and subsequently nearly all biomolecules to set up specific traits. When "unzipped", strands of DNA react with extra nucleotides to produce two identical double-paired strands, a process exploited in the Polymerase Chain Reaction, where a certain sequence of DNA is amplified using boundary primers and base-building polymerase.
At times, however, it may be useful to halt the replication of DNA, preserving a specific sample for analysis. Scientists are interested in mapping out the human genome and determining where specific nuclear proteins interact with the DNA chain[1]. In order to prevent DNA from replicating, strands may be cross-linked, or covalently bonded, with certain molecules. The network of cross-links essentially prevents the DNA from changing, but also causes it to die after a time. Because of this, the cross-linking process should be easily reversible in order to allow the DNA molecule to survive.
Cross-linking reagents must also exhibit stability under biomolecular analysis and be able to localize linking in a relatively precise area. One common cross-linking agent is formaldehyde, the simplest aldehyde. Formaldehyde's small size and durability under analytical conditions not normally found in the human body make it an ideal candidate[2]. In addition, incubation at 70oC undoes the cross-linking. Larger aldehydes fund in tobacco smoke and automotive exhaust can have similar but irreversible effects, permanently damaging DNA.
An example of where formaldehyde can be useful in cross-linking is the process of chromatin immunoprecipitation. This process analyses chromatin, which is the combination of DNA and proteins that comprises chromosomes. By mapping the locations of certain molecules known as histones, scientists can construct a crude map of proteins in the genome. First, however, the proteins and DNA need to be held together in place. The addition of formaldehyde fixes protein-protein and protein-DNA interactions, allowing analysts to survey the genome and then split it apart with particular antibodies and sonication, in the immunoprecipitation process. This isolates cross-linked sections of DNA from desired unbound sections, which scientists can amplify through PCR[3].
In some cases, scientists may want to observe the effects on DNA when these bacteria cells are oxidized. Too much of an oxidizing agent, such as H2O2 will kill the DNA, but too little will not illicit the response we want. To remedy this, cross-linking can be employed to guard the DNA against a high level of oxidzation. Disulfide bonds, formed by two sulfur molecules, are common bonds made to achieve this effect.
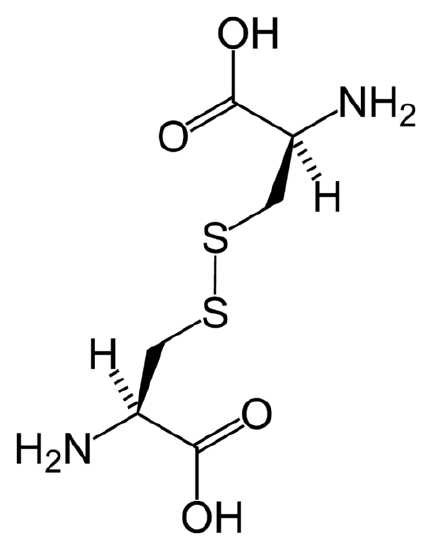
You can see in the above image of the protein cystine that the individual molecules each sulfur atom bonds seem identical. In fact, they are, and are called cysteine molecules; the disulfide bond unites two momomers to create a dimer, cystine. Disulfide bonds and other cross-linking agents are common reagents that produce dimers and polymers out of individual units.
In proteins like cystine, the addition of a sulfide bond can link together not just monomers, but entire lengths of protein to form secondary protein structure, like beta sheets. Sulfide bonds are critical in preventing polar water molecules from breaking apart the amide bonds.
Example \(\PageIndex{1}\): Bond Length
Based on what you know about trends in atomic sizes, determine which bond is longer: A disulfide bond or a C-C bond. From this, estimate which bond is stronger and whether a carbon backbone or disulfide bond would be more likely to break first.
Answer
Atomic radii increase moving down the periodic table due to an increasing number of orbitals, so the bond between two sulfur atoms would be longer than one between two carbon atoms. This means the disulfide bond would also be weaker than a C-C bond, and would probably break first.
Disulfide bonds are also more polarizable than carbon-carbon bonds, due to having a larger size and number of electrons. Electrophiles such as the halogens readily accept the electron pairs on the sulfur atoms, breaking the bond apart. A common electrophile is isothiocyanate, with functional group N=C=O[4]. You may see how the carbon atom can break a bond with the neighboring nitrogen and oxygen atoms in order to form two new single bonds with available pairs of electrons, such as those on the divalent sulfur atoms.
Disulfide bonds can play an important role in stabilizing the tertiary structure of proteins, lowering the entropy of the three-dimensional structure.
From ChemPRIME: 8.25: Cross-Linking
References
- ↑ www.sciencedirect.com/science...c2e20ecefa2d01
- ↑ www.sciencedirect.com/science.../sdarticle.pdf
- ↑ http://www.cancerepigenetics.com/tecniqueseng.htm
- ↑ www.journalarchive.jst.go.jp/...om=jnlabstract
Contributors and Attributions
Ed Vitz (Kutztown University), John W. Moore (UW-Madison), Justin Shorb (Hope College), Xavier Prat-Resina (University of Minnesota Rochester), Tim Wendorff, and Adam Hahn.


