Cisplatin 14. DNA
- Page ID
- 2911
Deoxyribonucleic acid, or DNA, stores all of our genetic information. It makes up the genes of all cells (animal and plant) as well as many viruses. In this module, we will examine the structure of DNA so that we can better understand its interaction with cisplatin. We will also examine replication and transcription, which are the processes by which genetic information is expressed. Finally, we will describe one mechanism by which damaged DNA is repaired in the cell.
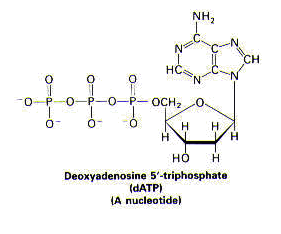
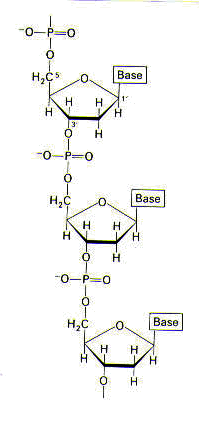
< face="Arial">Structure1 DNA is a remarkably long, threadlike macromolecule. DNA varies in length, depending on the organism. Certain viral DNA is 1.7 mm, whereas human DNA is nearly a meter in length. DNA is made up of many deoxyribonucleotide subunits. A nucleotide comprises a nitrogenous base, a sugar, and one or more phosphate groups, as shown below.
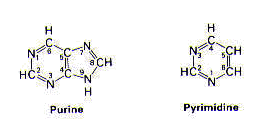
The nitrogenous base is a derivative of either purine or pyrimidine.
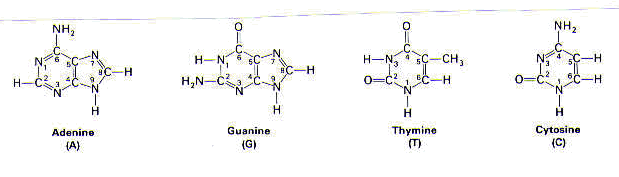
In DNA, the purines are adenine (A) and guanine (G), and the pyrimidines are thymine (T) and cytosine (C).
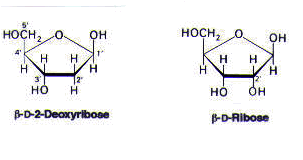
The sugar present in a deoxyribonucleotide is deoxyribose, which lacks an oxygen that is present in its parent compound, ribose.
The sugars and phosphate groups make up the backbone of DNA and therefore provide DNA’s structure; the backbone of DNA does not vary. The sequence of the bases does vary, however, and this is how genetic information is carried. The structure of part of a DNA chain is shown below.
The three-dimensional structure of DNA was deduced in 1953 by James Watson and Francis Crick. Their model of DNA had five important features: 1. Two helical chains of polynucleotides are coiled around a common axis; the chains run in opposite directions, as shown in Figure 1.
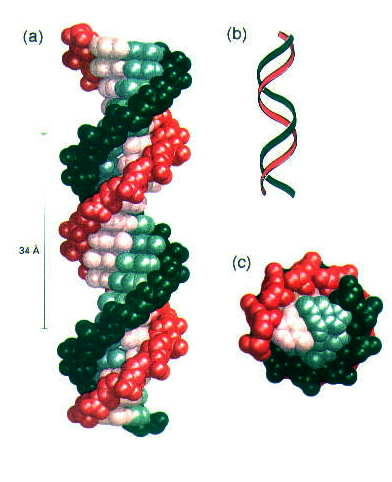
Figure 1. Model of a double-helical DNA. One chain is shown in green and the other in red. The purine and pyrimidine bases are shown in lighter colors than the sugar-phosphate backbone. (a) Axial view. The structure repeats along the helix axis (vertical) at intervals of 34 Å, which corresponds to 10 residues on each chain. (b) A schematic "ribbon" representation of an axial view of DNA. (Courtesy of M. Meselson and F. W. Stahl) (c) Radial view, looking down the helix axis. Reprinted with permission.1
2. The bases are on the inside of the helix, whereas the backbone (the sugars and phosphates) are on the outside. The bases are perpendicular to the helix axis and are said to stack (as plates are stacked when they are stored). The sugars are nearly parallel to the helix axis. See Figure 2.

Figure 2. Diagram of one of the strands of a DNA double helix, viewed down the helix axis. the bases (all pyrimidines here) are inside, whereas the sugar-phosphate backbone is outside. The bases are shown in blue and the sugars in red. Reprinted with permission.1
3. The diameter of the helix is 20 Å. The helical structure repeats after ten deoxyribonucleotide units on each chain (at intervals of 34 Å), as shown in Figure 1. 4. The two chains are held together by hydrogen bonds between pairs of bases. Adenine and thymine are always paired (and are joined by two hydrogen bonds); guanine and cytosine are always paired (and are joined by three hydrogen bonds), as shown in Figure 3.
 |
 |
Figure 3. Hydrogen bonding interactions (a) between adenine and thymine, and (b) between guanine and cytosine. Reprinted with permission.1
5. There are no restrictions on the sequence of bases along a polynucleotide chain, however, the precise sequence of bases carries the genetic information. < face="Arial">Replication1DNA duplicates itself by a process called replication. In replication, a strand from the original DNA molecule acts as a template for the formation of a new strand. The template can be either single- or double-stranded DNA; single-stranded DNA is formed when the hydrogen bonds between base pairs are broken. More than 20 proteins are required for the process of DNA replication. One of these is DNA polymerase I, which catalyzes the step-by-step addition of activated deoxyribonucleotide units to a DNA chain; the activated deoxyribonucleotide units for adenine, guanine, thymine, and cytosine are represented as dATP, dGTP, dTTP, and dCTP, respectively. Divalent magnesium (Mg2+) is also required for replication to occur. A schematic representation of DNA replication is shown in Figure 4.
Figure 4. A schematic representation of DNA replication, showing the addition of a guanine residue first, and then a thymine residue. Reprinted with permission.1
In short, replication describes the process by which genetic information is stored and transmitted. < face="Arial">Transcription< face="Arial">1 Genetic information is expressed through the processes of transcription, in which RNA (ribonucleic acid) is synthesized from a DNA template strand, and translation, in which proteins are synthesized from RNA—specifically, messenger RNA—templates. We will briefly discuss transcription here. Like DNA, RNA is made up of a sugar-phosphate backbone having four possible nitrogenous bases. Unlike DNA, however, ribose (shown below)—not deoxyribose—is found in RNA. The four nitrogenous bases in RNA are adenine, guanine, cytosine, and uracil (instead of thymine), as shown below.
An enzyme called RNA polymerase catalyzes the initiation and elongation of RNA chains, in which one activated ribonucleotide unit (represented by ATP, GTP, UTP, and CTP) is added at a time. The preferred template for transcription is double-stranded DNA. A divalent metal ion—either Mg2+ or Mn2+—is required for transcription. In short, transcription describes the first step in the expression of the genetic information stored by DNA. < face="Arial">Repair< face="Arial">1,2 DNA is damaged in a variety of ways, and all cells have mechanisms by which to repair the damage. DNA repair is possible because genetic information is stored in both strands of the double helix; therefore, information lost from one strand can be retrieved from the other. One way in which DNA becomes damaged is by the covalent cross-linking of DNA bases; cross-links can be formed from bases on either the same strand of the helix (intrastrand cross-linking) or opposite strands of the helix (interstrand cross-linking). Covalent intrastrand cross-links can be formed by chemical agents, such as cisplatin, or by ultraviolet light, leading to the formation of a pyrimidine dimer, as shown in Figure 5.
Figure 5. A thymine dimer, which is an example of a pyrimidine dimer. Reprinted with permission.1

Such a pyrimidine dimer cannot fit into the double helix, so the normal functions of the cell (such as replication and transcription) are blocked until the dimer is removed. The removal, or excision, of the dimer and the subsequent repair of the damaged DNA strand is referred to as the excision repair system and requires the action of several enzymes, three of which are described here and shown in Figure 6. After recognition of DNA damage by a damage recognition protein, the DNA strand containing the dimer is cut at two sites by an excinuclease. Then, the cut DNA strand is regenerated by DNA polymerase I. Finally, the newly replicated portion of the DNA strand is joined to the undamaged part of the DNA strand by DNA ligase.
Figure 6. Repair of a region of DNA containing a thymine dimer by the sequential action of a specific excinuclease, a DNA polymerase, and a DNA ligase. The thymine dimer is shown in blue, and the new region of DNA in red. (Courtesy of P. C. Hanawalt) Reprinted with permission.1
As we have seen from studies into the mode of action of cisplatin, covalent intrastrand cross-links can also be formed by chemical agents, such as cisplatin. A similar mechanism is believed to repair damage done to cancer cells by cisplatin, as we will see when we discuss drug resistance.
- (1) Stryer, L. Biochemistry, 4th ed. W. H. Freeman and Company: New York, 1995, Chapters 4, 5, and 31.
- (2) Pil, P., Lippard, S. J. In Encyclopedia of Cancer, J. R. Bertino, Ed. Academic Press: San Diego, CA, 1997, Vol. 1, pp. 392-410.