Cisplatin 13. Further Studies/Recent Developments
- Page ID
- 2910
The publication of the original papers in the 1960s describing the ability of certain platinum-containing compounds— specifically, cisplatin—to kill cancer cells has generated a tremendous amount of research. It would be impossible to describe all of that research here, so we will focus on just two areas of continuing study: the development of second- and third-generation drugs in the cisplatin family, and the research on the cellular target of cisplatin.
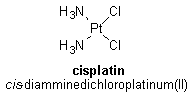
Second- and Third-Generation Drugs Once a successful drug has been discovered for the treatment of a particular disease or condition, researchers will often try to improve on that drug by synthesizing and studying related compounds, referred to as analogs or second-generation drugs. There are several reasons for making such analogs. First, the analogs may be able to improve on the efficacy of the original drug, meaning that lower doses are able to produce the same beneficial effects. Second, the toxicity profile of the analogs may be better than that of the original drug; in other words, the analogs may have fewer toxic side effects than the original drug. Third, the analogs may be able to be used to treat cases that have become resistant to the original drug. Finally, if the original drug can only be administered intravenously, the analogs may be able to be taken orally. After the efficacy and toxicity profiles of second-generation analogs are obtained, more analogs can be synthesized and studied; these analogs are called third-generation drugs. Many second-generation analogs of cisplatin have been made; some have been found to produce the same therapeutic effects as cisplatin—but with lower required doses and reduced side effects. Three of these analogs are carboplatin, spiroplatin, and iproplatin, shown below.
Carboplatin has proven to be the most useful of these three analogs, and was approved by the FDA for the treatment of ovarian cancers in 1989. Carboplatin and cisplatin have been shown to form an identical type of adduct with DNA and have similar activities against ovarian and lung tumors; however, carboplatin is less toxic to the peripheral nervous system and the kidneys. The lower toxicity of carboplatin compared to cisplatin is believed to be due to the structure of carboplatin. The presence of the bidentate dicarboxylate ligand (shown in Figure 1) in carboplatin slows down the degradation of carboplatin into potentially damaging derivatives.
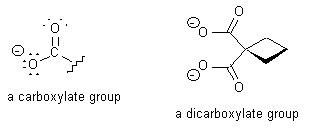
Figure 1. A carboxylate and a dicarboxylate group. Note that for simplicity, in the drawing of the dicarboxylate group, the electrons are not included, and the carbon atoms are not drawn out explicitly but rather are represented by the vertices in the structure. The dicarboxylate group is a bidentate ligand, meaning that it can bind a metal ion in two places.
Indeed, at 98.6 °F (37 °C) the retention half-life of carboplatin in blood plasma is 30 hours, whereas that for cisplatin is only 1.5-3.6 hours. In addition to the lower toxicity of carboplatin, it has been shown to work in some cases when cisplatin has failed.1,2 The decreased toxicity of carboplatin and the activity of carboplatin against cisplatin-resistant tumors have led to greater use of carboplatin which has resulted in carboplatin becoming the greater money-maker of the two drugs. (see module on profits) In addition to carboplatin and other second-generation cisplatin analogs, several third-generation drugs have been synthesized and tested. One broad class of these drugs, having the general structure shown in Figure 2, can be taken orally—a significant improvement over cisplatin, which must be administered intravenously.
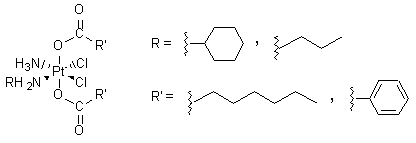
Figure 2. The general structure of ammine/amine platinum(IV) dicarboxylates. Many different R and R’ groups were tried; two examples for each are shown here.
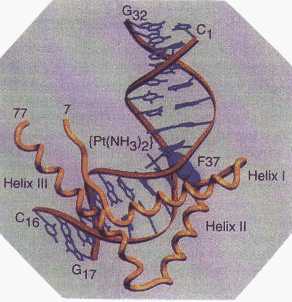
These complexes are stable enough to survive passage down the digestive tract. They are then transported across the gastrointestinal mucosa into the bloodstream. After absorption into the bloodstream, these compounds are metabolized to form four-coordinate, platinum(II) cisplatin analogs. These new four-coordinate complexes are assumed to be the active forms of the drug.1,3 A variety of six-coordinate platinum(IV) complexes were tested in cell culture using six human ovarian cancer cell lines. Researchers found that as the ring size of ligand R increased, the complexes became increasingly more effective than cisplatin at killing cancer cells. Moreover, like carboplatin, these complexes were also toxic to certain cancer cell lines that were resistant to cisplatin. Researchers believe that these third-generation drugs are more effective than cisplatin because they are taken up into the cell in greater concentrations than cisplatin.3 Cellular Target of Cisplatin The cellular target of cisplatin has long been believed to be DNA (see modules on mode of action and DNA). Cisplatin has been shown to bind DNA and cause the DNA duplex to bend and unwind. For many years, however, researchers have been trying to determine precisely how the cisplatin–DNA complex leads to cell death. A recent crystal structure4 provides some clues about what may be occurring on a subcellular level. The research group of Stephen Lippard at the Massachusetts Institute of Technology has crystallized a ternary complex (that is, one having three components) comprising cisplatin, DNA, and a cellular protein called a high mobility group (HMG) protein. This protein inserts a phenyl group protruding from its backbone5 into the notch created when cisplatin forms a complex with DNA and causes it to bend, as shown in Figure 3. A mutant protein lacking this phenyl group does not form a complex with cisplatin and DNA, suggesting that the phenyl group is crucial for complex formation. The interaction of an HMG-protein with the cisplatin–DNA complex may shield DNA from intracellular repair; the inability of DNA to repair itself may cause cancer cells to die. (See module on mode of action) Knowledge of this ternary complex should aid in the development of future anticancer strategies. Scientists may now use the information about HMG-proteins to design proteins that will bind the cisplatin–DNA complex even more tightly. In addition, they may use the knowledge gained from this research to continue to look for cisplatin analogs that will form even better drug–DNA–protein complexes.6
- Bertini, I., Gray, H. B., Lippard, S. J., Valentine, J. S. Bioinorganic Chemistry. University Science Books: Sausalito, CA, 1994.
- Kaim, W., Schwederski, B. Bioinorganic Chemistry: Inorganic Elements in the Chemistry of Life. John Wiley & Sons Ltd: Chichester, England, 1994.
- Kelland, L. R., Murrer, B. A., Abel, G., Giandomenico, C. M., Mistry, P., Harrap, K. R. Cancer Research ,1992, 52, pp. 822-828.
- A crystal structure shows the three-dimensional arrangement of atoms in a molecule and is obtained from the diffraction pattern of x-rays passing through a crystal of the molecule.
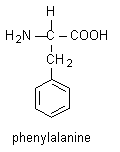
- The phenyl group is the side chain of phenylalanine, which is an amino acid contained in the backbone of the HMG-protein. There are 20 naturally occurring amino acids; these are the building blocks of proteins.
- Ohndorf, U.-M., Rould, M. A., He, Q., Pabo, C. O., Lippard, S. J. Nature, 1999, 399, pp. 708-712.


