Cytochrome C
- Page ID
- 97954
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)All cells require energy in the form of ATP, Adenosine Triphosphate, to drive essential metabolic processes for survival. When an aerobic organism digests its food, glucose (C6H12O6) is broken down into two molecules of pyruvate in the process of glycolysis. The reduced electron carriers, NADH and FADH2 are produced as a byproduct of this reaction.
NAD+ gains two electrons (2e-) and a hydrogen ion (H+) to form NADH. Similarly, FAD accepts two hydrogen ions (2H+) and two electrons (2e-) to form FADH2. These electrons from NADH and FADH2 then enter the complexes of the Electron Transport Chain. The Electron Transport Chain (ETC) is a series of electron transport proteins located in the inner membrane of the mitochondria-- the powerhouse of the cell (Figure 1). Embedded in the mitochondrial membrane is a series of redox-active proteins, which act like a wire by shuttling electrons through the ETC. Through a series of redox reactions, these proteins pump protons (H+) from the mitochondrial matrix to the intermembrane space. The accumulation of protons in the intermembrane space generates an electrochemical gradient which powers the synthesis of ATP by the enzyme ATP Synthase. The Electron Transport Chain produces a total of 34 molecules of ATP which the cell can use to carry out its metabolic processes for survival. 1
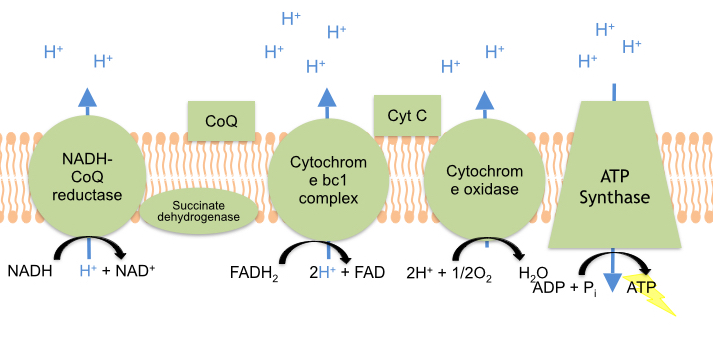
This is a general overview of The Electron Transport Chain (ETC). The ETC is a series of four protein complexes: NADH dehydrogenase, succinate dehydrogenase, cytochrome bc1, and cytochrome c oxidase, embedded in the inner membrane of the mitochondria. As electrons are transferred through these protein complexes, a proton (H+) gradient accumulates in the intermembrane space of the mitochondria. ATP Synthase uses this proton gradient established by the ETC to synthesize energy in the form of ATP.
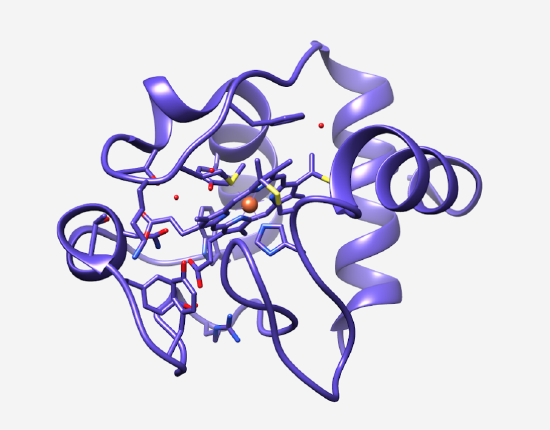 |
Cytochrome c is an important component of the Electron Transport Chain for the synthesis of ATP (Figure 2). Cytochrome c is a water soluble electron transport protein that is loosely associated with the mitochondrial inner membrane. In the Electron Transport Chain, cytochrome c transfers one electron at a time via its heme group from the third complex of the electron transport chain, cytochrome bc1, to the fourth complex of the electron transport chain, cytochrome c oxidase.2 Cytochrome c contains a heme iron metal center that is essential to its function. During the electron transport process, this heme iron interconverts between the Fe3+ and Fe2+ oxidation states, which allows for electrons to be accepted and donated.4 When cytochrome c is in its oxidized form, an electron is transferred from the cytochrome bc1 complex to the heme Fe3+, reducing it to Fe2+. Finally, cytochrome c releases the electron to the final electron carrier of the ETC, cytochrome c oxidase. At this point, the iron center will return to its Fe3+oxidation state. |
The iron metal center of cytochrome c represents an octahedral geometry due to the coordination of six ligands around the central iron ion (Figure 3). An octahedral geometry is preferred for cytochrome c because each of the 6 electron rich ligands contribute to stabilizing the positively charged metal iron ion. The heme iron shown in orange is coordinated by four nitrogen atoms of a rigid square planar porphyrin ring and two axial ligands: the sulfur atom of a methionine residue and a nitrogen atom of a histidine imidazole ring. The porphyrin ring of cytochrome c is considered to be a tetradentate chelating ligand because the four nitrogen atoms of the porphyrin ring bind to the central iron, forming a stable organometallic complex. Based on the chelate effect, this tetradentate ligand binding site is more entropically favorable compared to the affinity of a monodentate ligand for the same metal ion. 5
The ligands of cytochrome c are appropriate based on Hard Soft Acid Base Theory. (HSAB). HSAB categorizes acids and bases as hard, soft, or borderline. Soft acids and bases are larger and easily polarizable while hard acids and bases are smaller and less polarizable. Species that lie in between hard and soft are considered borderline. 6 Based on HSAB, it is logical that the borderline iron coordinates with the nitrogen of the histidine imidazole ring (a borderline base), the nitrogen atoms provided by the porphyrin ring (borderline bases), and the sulfur of the methionine (a soft base).
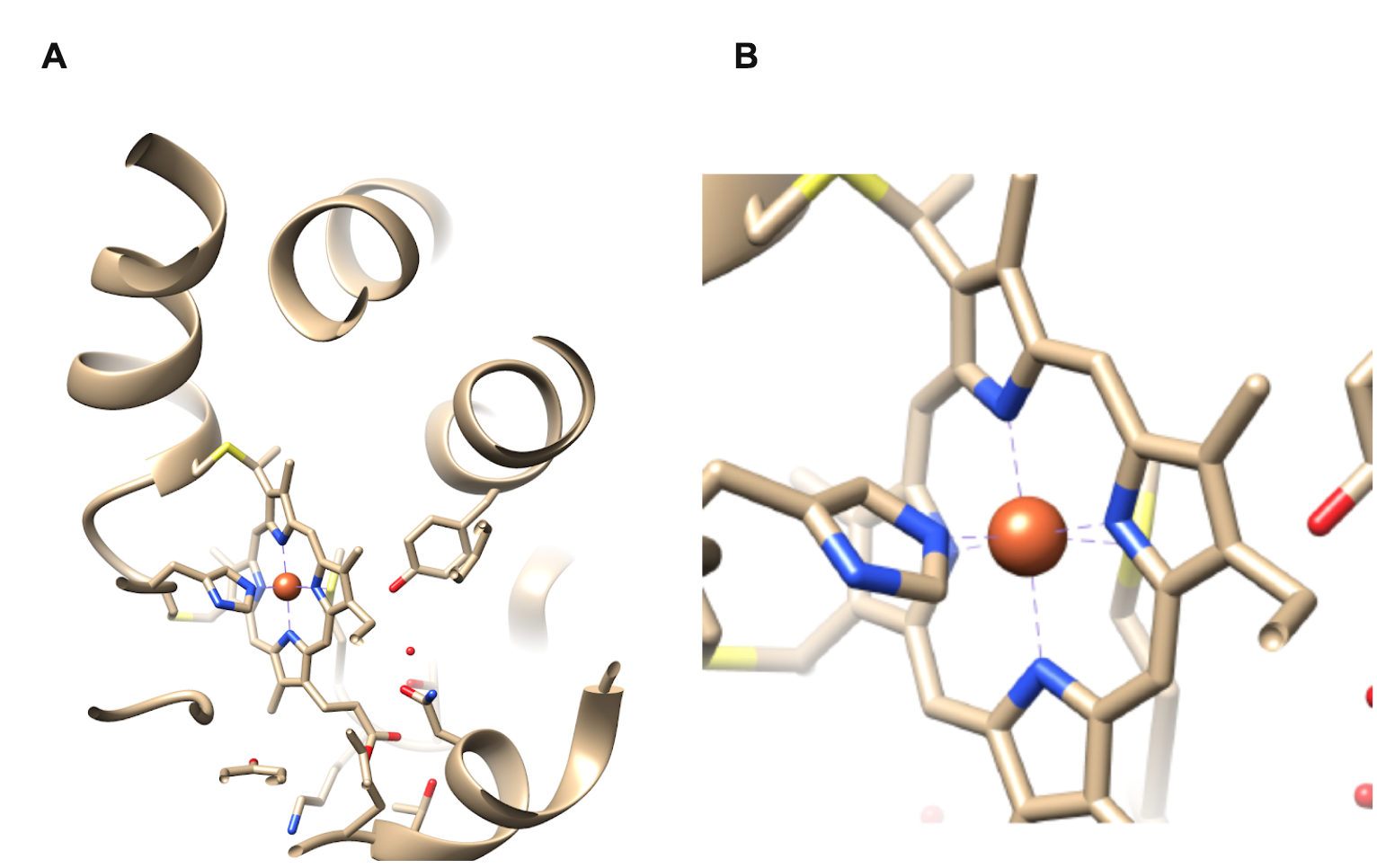
Figure 3A shows the entire structure of cytochrome c while Figure 3B is a magnified inset of the heme iron metal center essential to the electron transport function of cytochrome c. The heme iron shown in orange is coordinated by four nitrogen atoms of a rigid square planar porphyrin ring (blue) as well as two axial ligands: a sulfur atom of a methionine residue (yellow) and a nitrogen atom of a histidine imidazole ring (blue).
The iron metal center of cytochrome c represents an octahedral geometry due to the coordination of six ligands around the central iron ion (Figure 3). An octahedral geometry is preferred for cytochrome c because each of the 6 electron rich ligands contribute to stabilizing the positively charged metal iron ion. The heme iron shown in orange is coordinated by four nitrogen atoms of a rigid square planar porphyrin ring and two axial ligands: the sulfur atom of a methionine residue and a nitrogen atom of a histidine imidazole ring. The porphyrin ring of cytochrome c is considered to be a tetradentate chelating ligand because the four nitrogen atoms of the porphyrin ring bind to the central iron, forming a stable organometallic complex. Based on the chelate effect, this tetradentate ligand binding site is more entropically favorable compared to the affinity of a monodentate ligand for the same metal ion.5
The ligands of cytochrome c are appropriate based on Hard Soft Acid Base Theory. (HSAB). HSAB categorizes acids and bases as hard, soft, or borderline. Soft acids and bases are larger and easily polarizable while hard acids and bases are smaller and less polarizable. Species that lie in between hard and soft are considered borderline. 6 Based on HSAB, it is logical that the borderline iron coordinates with the nitrogen of the histidine imidazole ring (a borderline base), the nitrogen atoms provided by the porphyrin ring (borderline bases), and the sulfur of the methionine (a soft base).
Although the heme iron metal center changes oxidation state during the electron transport proces, cytochrome c always adopts an octahedral, low spin geometry regardless of the oxidation state on the iron. 7 This geometry is preferred based on Ligand Field Stabilization Energy (LFSE). LFSE is the total energy of the d-electrons of a metal complex relative to the theoretical barycenter. The formula for determining LFSE is shown in Equation 1 below, where x= number of d-electrons in the low energy t2g orbitals and y= the number of d-electrons in the high energy eg orbitals.
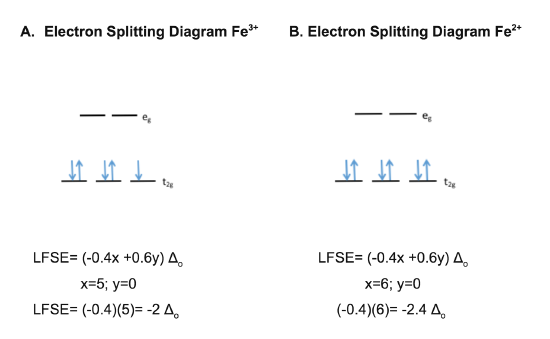
The electron splitting diagrams and calculations for low spin Fe3+ and Fe2+ are shown in figures A and B, respectively. The more negative LFSE values for low spin iron compared to high spin iron indicate that this low spin conformation is more energetically favorable and is therefore the configuration that the complex will adopt.

The theoretical electron splitting diagrams and calculations for high spin Fe3+ and Fe2+ are shown in figures A and B, respectively. However, the LFSE values are more negative for low spin iron than for high spin iron, regardless of the oxidation state. Therefore, the heme iron metal center of cytochrome c will always adopt a low spin octahedral geometry regardless of the oxidation state on the iron.
In the low spin state, the d-electrons pair in the low energy t2g orbitals before occupying the higher energy eg orbitals, which increases stability. In the high spin state, the d-electrons singly occupy both the t2g and eg orbitals, without regard to the energy of the orbitals, and then pair up. The more negative the LFSE, the more stable the complex. The LFSE values for low spin iron are more negative than the LFSE values for high spin iron, regardless of the oxidation state present on the iron. Thus, the iron metal center of cytochrome c will always adopt the more energetically favorable low spin conformation.
During the electron transport process, the heme iron of cytochrome c cycles between the +2 and +3 oxidation states. As cytochrome c accepts an electron from the third complex of the electron transport chain, cytochrome bc1, the Fe3+ iron metal center is reduced to Fe2+. When cytochrome c releases this electron to the final complex of the electron transport chain, cytochrome c oxidase, the Fe3+oxidation state is restored. However, despite this change in the oxidation state of the iron, the heme coordination does not change; the iron ion is “locked in place” and undergoes minimum reorganization.
Reduction potential also facilitates the electron transport function of cytochrome c in the ETC. Reduction potential (Eo’) is the tendency for a chemical species to acquire electrons and therefore be reduced. The more positive the reduction potential, the greater the tendency for that chemical species to accept an electron and to be reduced. The reduction potential for cytochrome bc1 (complex III of the ETC) is 0.194 V. Cytochrome bc1 donates one electron to the oxidized form of cytochrome c (Fe3+), reducing the iron of cytochrome c by one oxidation state to become Fe2+. The reduction potential for cytochrome c is 0.254 V. The reduction potential for cytochrome c oxidase (Complex IV of the ETC) is 0.562 V. 8 Cytochrome C Oxidase accepts an electron from the reduced cytochrome c (Fe2+) returning cytochrome c to its oxidized form (Fe3+). The complexes of the Electron Transport Chain are arranged in order of increasing redox potential (each complex has higher affinity for electrons than the previous), which drives the flow of electrons towards the final complex of the Electron Transport Chain, Cytochrome C Oxidase.
Eo’donor < Eo’ electron transfer protein/center < Eo’acceptor: Must be true for reaction to be spontaneous!
In conclusion, cytochrome c is an essential electron transfer protein which shuttles electrons between complexes III and IV of the ETC. The heme iron metal center readily interconverts between the Fe3+ and Fe2+ which allows for electrons to be accepted and donated. If this electron transfer did not occur, the ATP required to power many metabolic processes such as muscle movement, DNA synthesis, and active transport would not be produced. Therefore, cytochrome c is an important component in facilitating the production of this ATP through its electron transfer to Cytochrome C Oxidase, which provides cells the energy they need to carry out these vital processes necessary for survival.9
Sources
- Alberts, B.; Johnson, A.; Lewis, J, et al. Molecular Biology of the Cell. 4th ed.; New York. Garland Science; 2002. Electron Transport Chains and Their Protein Pumps. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26904/
- Lehninger, A.L.; Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry; Macmillan Higher Education: Houndmills, 2017.
- Takano, T.; Dickerson, R.E. Redox conformation changes in refined tuna cytochrome c. Proc. Natl. Acad. Sci. USA. 1980. 77, 6371-6375.
- Roat-Malone, R.M. Bioinorganic Chemistry- a short course; Wiley & Sons: Hoboken, NJ, 2007.
- Xinshan, K. & Carey, J. (1999). Role of Heme in Structural Organization of Cytochrome c Probed by Semisynthesis. Biochemistry, 38 (48), 15944-15951.
- Chemistry LibreTexts: Hard and Soft Acids and Bases
- Silva, J.J.R.; Williams, R.J.P. The biological chemistry of the elements: the inorganic chemistry of life; Oxford University Press: Oxford, 2001.
- Bertini, I. Biological inorganic chemistry: structure and reactivity; University Science Books: Sausalito, CA, 2007.
- Erecińska, M.; Silver, I.A. ATP and Brain Function. Journal of Cerebral Blood Flow and Metabolism. 1989, 9, 2-19.
Contributed By
- This work was originally written by Hannah Voss, Spring 2018: Hannah is currently (as of 2018) a junior biology major at Saint Mary's College in Notre Dame, IN.
- This work was originally edited by Dr. Kathryn Haas (Assistant Professor) and Madison Sendzik (Teaching and Research Assistant) at Saint Mary's College.


