CX11. Protein Modifications
- Page ID
- 4242
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)After proteins are synthesized via transcription of DNA, their structures may be modified in a number of ways. This process, in general, is referred to as "post-translational modification." It might involve the attachment of small organic groups to the protein, attachment of other biomolecules, or even the complete alteration of an amino acid residue into another form, so that the protein actually contains a residue that does not exist in the basic beginner's amino acid alphabet.
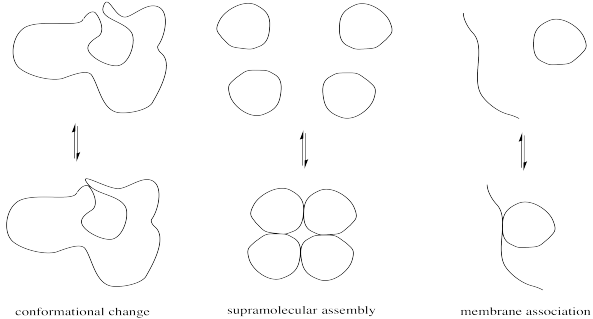
Minor modifications of proteins can be very important in regulating enzyme activity. Modest changes in structure can have a big impact on the conformation of the protein, which in turn might open or close access into the enzyme's interior or alter the shape of the active site. Alternatively, these changes might affect the location of a protein by moving it from the cell membrane into the cell interior or vice versa. They might also result in the formation or dissociation of supramolecular assemblies, by increasing or decreasing the attraction between two separate biomolecules.
The physical reasons for these structure-property changes are based in simple intermolecular attractions or, in the case of a large, complicated protein, intramolecular attractions between different parts of the same protein. A hydrogen bonding interaction or ion-dipole interaction that holds the protein in one conformation may be disrupted if a key player in that interaction is suddenly masked. For example, a cationic ammonium side chain may react to become an amide group; the absence of a full positive charge on this group significantly alters its intermolecular attractions.
There are probably hundreds of ways in which proteins are modified. We will look at just a few different modifications that occur in proteins, some of which are closly tied to carboxylate substitution. Note that any of these modifications might act to turn an enzyme "off" or "on"; the details depend on the individual case.
Acetylation
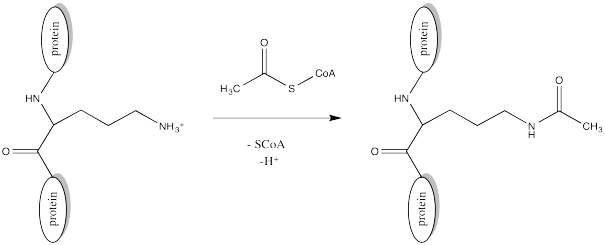
Acetylation is the attachment of an acetyl group, CH3C=O, to another compound such as an amino acid residue. Serine and threonine groups might be acetylated to make esters, cysteine side chains might be acetylated to make thioesters, or amino groups. Frequently, acetylation refers specifically to reaction at an amino group, such as the N-terminus of a protein or in a lysine side chain, forming an amide.
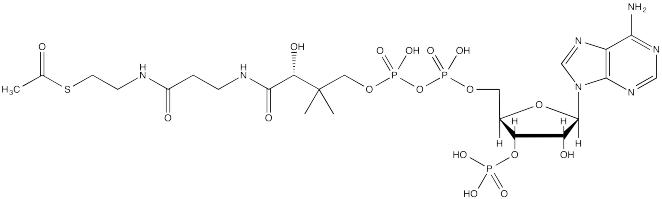
The source of the acetyl group is acetyl coenzyme A (AcSCoA, below), the thioester workhorse of the cell. This structure may seem a little bit complicated, but at the most basic level it is just a carboxyl electrophile (CH3C=O) attached to a very large thiolate leaving group.
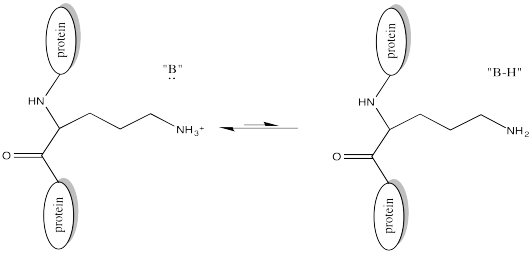
Of course, lysine side chains and the N-termini of proteins are usually in a protonated state under biological conditions. That means that there must be a deprotonation step along the reaction pathway.
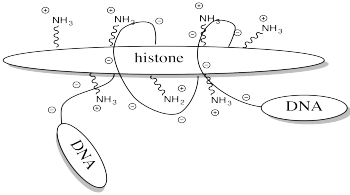
One example of the role of acetylation is seen in histones. Histones are proteins found in chromatin, the mixture of DNA and proteins in the cell nucleus. The already-coiled DNA helix is further wrapped around histones, bundling it up into a smaller package. DNA has many, many negative charges all along it because the phosphate units in its phosphate-sugar copolymer backbone are negatively charged at typical biological pH. The DNA interacts easily with the histones because they are rich in positively charged lysine residues.
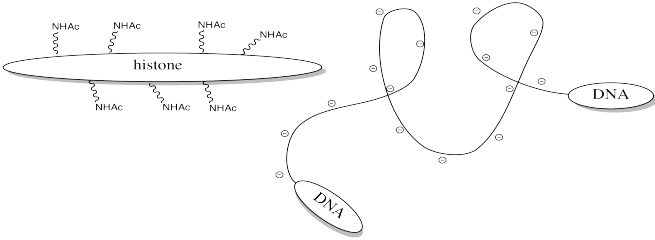
Storing DNA in smaller bundles lets you keep more junk in your nucleus, but you don't want to just let the DNA sit there. Once in a while you want to take it out and do something with it, but how can you do that when it's stuck to those darned histones? The answer is, you just have to turn off the histones' force field. Get rid of that positive charge, and the DNA won't be stuck anymore. It's easy to do that by acetylating the histones.
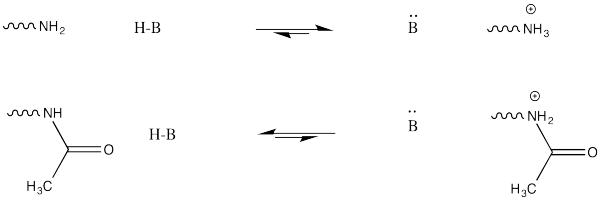
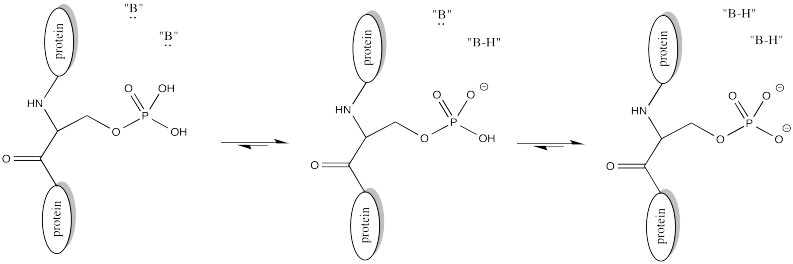
This change in charge results because, although amines are easily protonated, amides are not. Thus, amines are likely to carry positive charge under biological conditions, whereas amides are likely to be neutral.
Problem CX10.1.
Explain why an amine is easily protonated but an amide is not.
Problem CX10.2.
Provide a mechanism for the acetylation of a lysine side chain. Assume the presence of histidine and its conjugate acid, histidinium ion; these species are common proton shuttles in biological reactions.
Phosphorylation
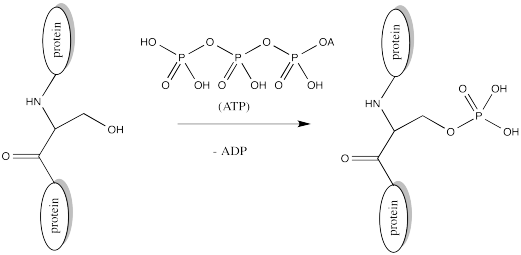
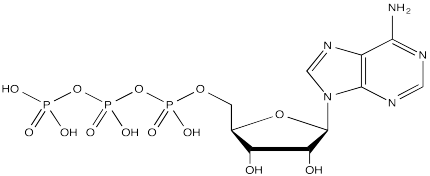
Serine, tyrosine and threonine residues are frequently modified by phosphorylation, which is the formation of a phosphate ester. The phosphorylation of these groups results in a change from a neutral side chain to an anionic side chain. The phosphate donor in these cases is another ubiquitous cellular performer, ATP.
The complete structure of ATP is shown here. It is a little less complicated than AcSCoA. Once again, at the most basic level of reactivity, it provides a phosphate electrophile, attached to a phosphate leaving group.
The anionic nature of the phosphate may not be apparent in the above drawing of ATP, but in reality, at biological pH, a phosphate would be deprotonated. Phosphates actually undergo multiple equilibria involving the loss of two protons.
Problem CX10.3.
Provide a mechanism for phosphorylation of serine in the presence of an appropriate biological proton shuttle.
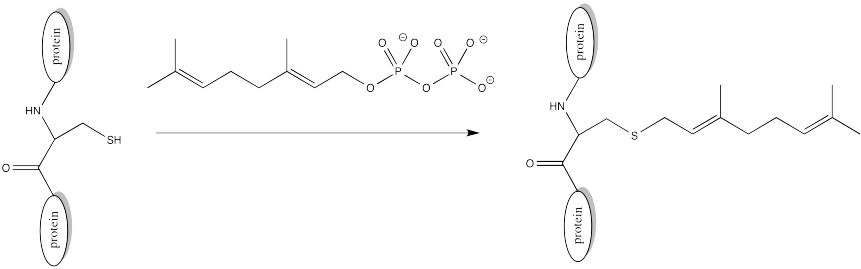
Alkylation (Farnesylation)
There are other modifications that occur via different mechanisms. In a farnesylation, reaction does not even take place at a carbonyl. It does not take place at a phosphonate or a sulfonate.
Problem CX10.4.
Sulfur is the nucleophile in the farnesylation shown above. Identify the electrophile and show a mechanism with curved arrows.
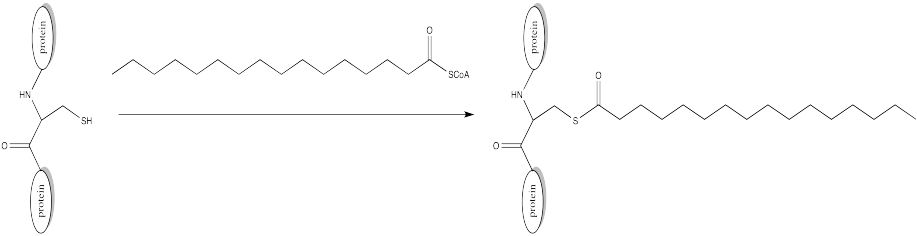
Problem CX10.5.
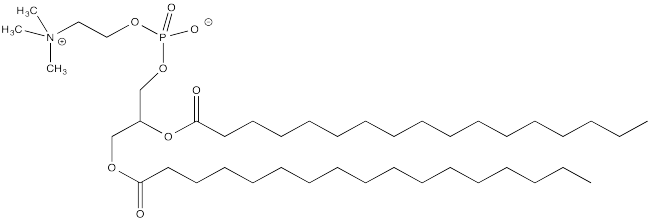
Palmitoylation is closely related to acetylation, but the electrophile in this case is a palmitoyl group instead of an acetyl.
- Provide a mechanism for palmitoylation of cysteine in the presence of an appropriate biological proton shuttle.
- Explain how palmitoylation might influence the interaction of a protein with a cell membrane, composed of phospholipids like this one: