5.7: Spin-Spin Splitting in Proton NMR Spectra
- Page ID
- 432195
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Objectives
After completing this section, you should be able to
- understand what spin-spin splitting is and what information it tells you
- explain the spin-spin splitting pattern observed in the 1H NMR spectrum of a simple organic compound.
- interpret the splitting pattern of a given 1H NMR spectrum.
- determine the structure of a relatively simple organic compound, given its 1H NMR spectrum and other relevant information.
- use coupling constants to determine which groups of protons are coupling with one another in a 1H NMR spectrum.
- predict the splitting pattern which should be observed in the 1H NMR spectrum of a given organic compound.
Key Terms
Make certain that you can define, and use in context, the key terms below.
- coupling constant
- multiplet
- quartet
- triplet
- doublet
From what we have learned about 1H NMR spectra so far, we might predict that the spectrum of 1-chloroethane, CH3CH2Cl, would consist of two peaks—one, at about 0.9 δ, expected for CH3 and one shifted downfield because of the presence of an additional electronegative chlorine atom on the second carbon. However, when we look at the spectrum (below) it appears to be much more complex. True, we see absorptions in the regions we predicted, but instead of the single peak we have seen thus far, these absorptions appear as a group of three peaks (a triplet) and a group of four peaks (a quartet). This complication is in fact very useful to the organic chemist, and adds greatly to the power of NMR spectroscopy as a tool for the elucidation of chemical structures. The split peaks (multiplets) arise because the magnetic field experienced by the protons of one group is influenced by the spin arrangements of the protons in an adjacent group.

The source of spin-spin coupling
The 1H-NMR spectra that we have seen so far (of methyl acetate and para-xylene) are somewhat unusual in the sense that in both of these molecules, each set of protons generates a single NMR signal. In fact, the 1H-NMR spectra of most organic molecules contain proton signals that are split into two or more sub-peaks. This splitting behavior actually provides us with more information about our sample molecule.
Consider the spectrum for 1,1,2-trichloroethane. In this and in many spectra to follow, enlargements of individual signals will be shown so that the signal splitting patterns are recognizable.
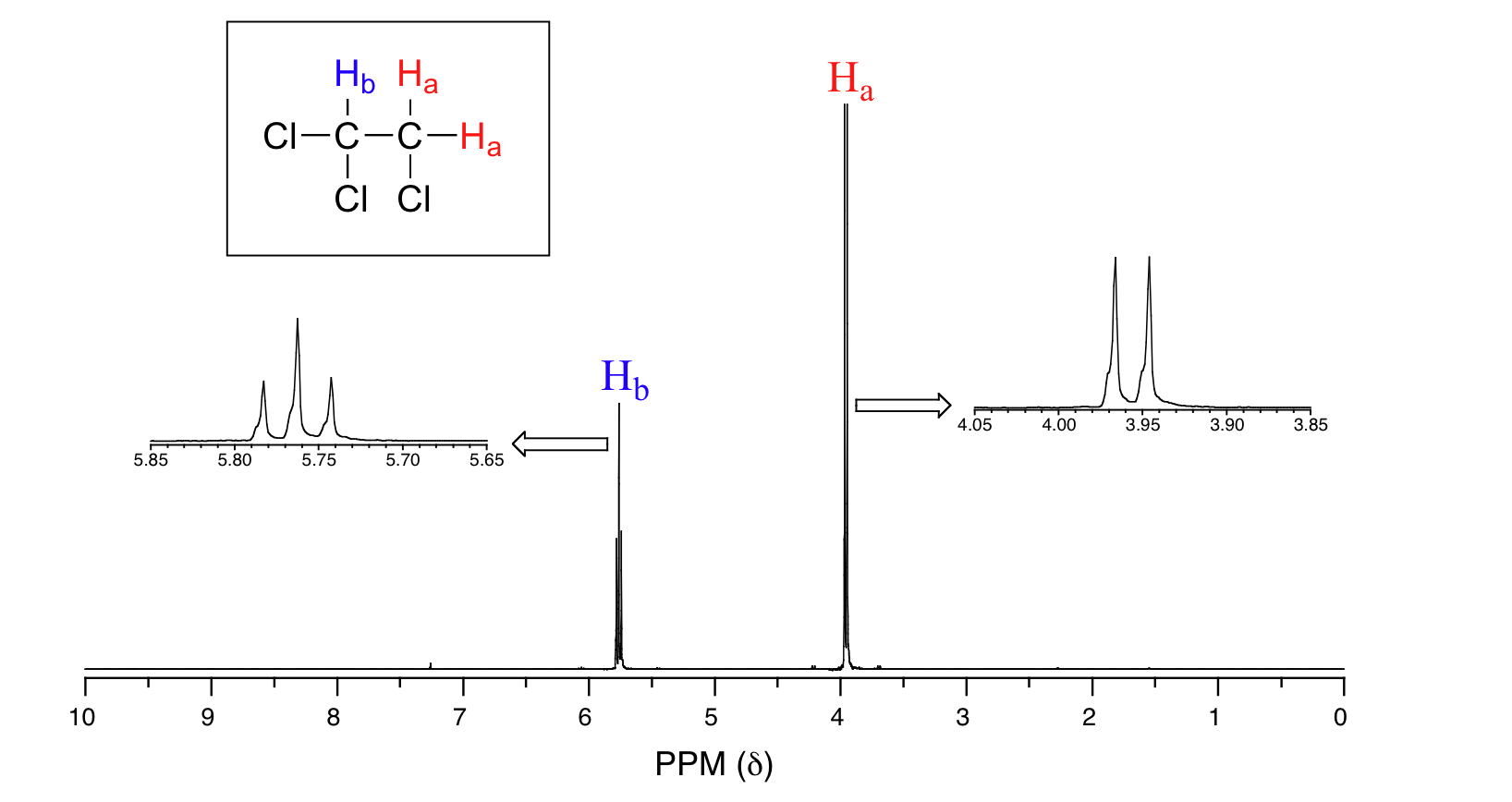
The signal at 3.96 ppm, corresponding to the two Ha protons, is split into two subpeaks of equal height (and area) – this is referred to as a doublet. The two Hb protons at 5.76 ppm, on the other hand, is split into three sub-peaks, with the middle peak higher than the two outside peaks - if we were to integrate each subpeak, we would see that the area under the middle peak is twice that of each of the outside peaks. This is called a triplet.
The source of signal splitting is a phenomenon called spin-spin coupling, a term that describes the magnetic interactions between neighboring, non-equivalent NMR-active nuclei. In our 1,1,2--trichloroethane example, the Ha and Hb protons are spin-coupled to each other. Here's how it works, looking first at the Hb signal. The magnetic environment experienced by Hb is influenced by the fields of both neighboring Ha protons, which we will call Ha1 and Ha2. There are four possibilities here, each of which is equally probable. First, the magnetic fields of both Ha1 and Ha2 could be aligned with B0, which would deshield Hb, shifting its NMR signal slightly downfield. Second, both the Ha1 and Ha2 magnetic fields could be aligned opposed to B0, which would shield Hb, shifting its resonance signal slightly upfield due to being more shielded. Third and fourth, Ha1 could be with B0 and Ha2 opposed, or Ha1 opposed to B0 and Ha2 with B0. In each of the last two cases, the shielding effect of one Ha proton would cancel the deshielding effect of the other, and the chemical shift of Hb would be unchanged. These ideas an be illustrated by a splitting diagram, as shown below.
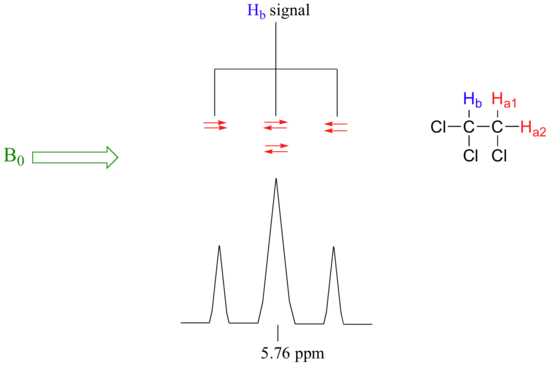
So in the end, the signal for Hb is a triplet, with the middle peak twice as large as the two outer peaks because there are two ways that Ha1 and Ha2 can cancel each other out.
The Ha hydrogens give rise to a doublet signal at 3.96 ppm – notice that the two peaks are equal in height to one another. This splitting pattern results from the spin-coupling effect of the one Hb hydrogen next door, and can be explained by an analysis similar to that which we used to explain the triplet pattern above. The proton could be aligned with the magnetic field, which would deshield Ha or it could oppose the magnetic field, which would shield Ha.
Now, consider the spectrum for ethyl acetate:

Explain the splitting patterns of Ha, Hb, and Hc in ethyl acetate.
Solution
We see an unsplit singlet peak at 1.83 ppm that corresponds to the acetyl (Ha) hydrogens. This signal is unsplit because there are no hydrogens on the adjacent carbon. The signal at 1.055 ppm for the Hc hydrogens is split into a triplet by the two Hb hydrogens next door. The explanation here is the same as the explanation for the triplet peak we saw previously for 1,1,2-trichloroethane.
The Hb hydrogens give rise to a quartet signal at 3.91 ppm – notice that the two middle peaks are taller then the two outside peaks. This splitting pattern results from the spin-coupling effect of the three Hc hydrogens next door, and can be explained by an analysis similar to that which we used to explain the doublet and triplet patterns previously discussed.
Instead of drawing a splitting diagram for every molecule, there is a recognizable pattern which is usually referred to as the n + 1 rule: if a set of hydrogens has n neighboring, non-equivalent hydrogens, it will be split into n + 1 subpeaks. This is very useful information if we are trying to determine the structure of an unknown molecule: if we see a triplet signal, we know that the corresponding hydrogen or set of hydrogens has two neighboring hydrogens. When we begin to determine structures of unknown compounds using 1H-NMR spectral data, it will become more apparent how this kind of information can be used.
Three important points need to be emphasized here. First, signal splitting only occurs between non-equivalent hydrogens – in other words, Ha1 in 1,1,2-trichloroethane is not split by Ha2, and vice-versa.
Second, splitting occurs primarily between hydrogens that are separated by three bonds. This is why the Ha hydrogens in ethyl acetate form a singlet– the nearest hydrogen neighbors are five bonds away (count the bonds between hydrogen to hydrogen), too far for coupling to occur.
Occasionally we will see four-bond and even 5-bond splitting, but in these cases the magnetic influence of one set of hydrogens on the other set is much more subtle than what we typically see in three-bond splitting (more complex coupling interactions is provided in section 5.8).
Finally, splitting is most noticeable with hydrogens bonded to carbon. Hydrogens that are bonded to heteroatoms (alcohol or amino hydrogens, for example) are coupled weakly - or not at all - to their neighbors. This has to do with the fact that these protons exchange rapidly with solvent or other sample molecules, which is greatly enhanced by even traces of acid or base (see mechanism with ethanol below). Remember, NMR is like a camera that takes photographs of a rapidly moving object with a slow shutter speed - the result is a blurred image. This blurred image here creates a broader singlet for exchanging protons, even if there are neighboring hydrogens.
To see the splitting of exchangeable protons, the NMR experiment needs to be carried out as low temperatures.
Multiplicity in Proton NMR
The number of lines in a peak is always one more (n+1) than the number of hydrogens on the neighboring carbon. This table summarizes coupling patterns that arise when protons have different numbers of neighbors.
|
# of lines |
ratio of lines |
term for peak |
# of neighbors |
|
1 |
- |
singlet |
0 |
|
2 |
1:1 |
doublet |
1 |
|
3 |
1:2:1 |
triplet |
2 |
|
4 |
1:3:3:1 |
quartet |
3 |
|
5 |
1:4:6:4:1 |
quintet |
4 |
|
6 |
1:5:10:10:5:1 |
sextet |
5 |
|
7 |
1:6:15:20:15:6:1 |
septet |
6 |
|
8 |
1:7:21:35:35:21:7:1 |
octet |
7 |
|
9 |
1:8:28:56:70:56:28:8:1 |
nonet |
8 |
Coupling constants
Chemists quantify the spin-spin coupling effect using something called the coupling constant, which is abbreviated with the capital letter J. The coupling constant is simply the difference, expressed in Hz (not ppm), between two adjacent sub-peaks in a split signal. For our triplet in the 1,1,2-trichloroethane spectrum, for example, the three subpeaks are separated by 6.1 Hz, and thus we write 3Ja-b = 6.1 Hz.
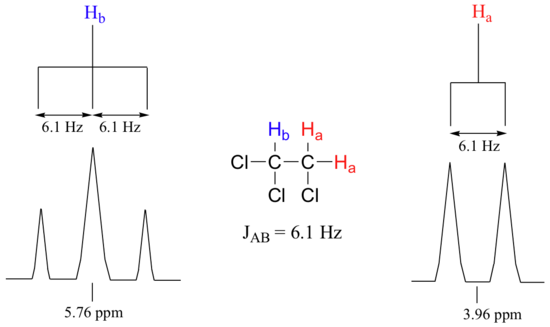
The superscript 3 tells us that this is a three-bond coupling interaction, and the a-b subscript tells us that we are talking about coupling between Ha and Hb. Unlike the chemical shift value, the coupling constant, expressed in Hz, is the same regardless of the applied field strength of the NMR magnet. This is because the strength of the magnetic moment of a neighboring proton, which is the source of the spin-spin coupling phenomenon, does not depend on the applied field strength. Also, the coupling constant 3Ja-b quantifies the magnetic interaction between the Ha and Hb hydrogen sets, and this interaction is of the same magnitude in either direction. In other words, Ha influences Hb to the same extent that Hb influences Ha. When looking at more complex NMR spectra (Section 5.8), this idea of reciprocal coupling constants can be very helpful in identifying the coupling relationships between proton sets.
Similar types of bonds lead to similar coupling constants and this has resulted in a range for particular types of hydrogens interacting through a particular bond. For example, coupling constants between proton sets on neighboring sp3-hybridized carbons (3 bonds) is typically in the region of 6-8 Hz. The table below lists typical three-bond coupling constant values for different types of bonds. Remember, chemically equivalent protons do not couple with one another to give spin-spin splitting.
| Type of bond | Image of type of bond | Coupling constant (Hz) |
| Standard three bond coupling | 6-8 | |
| Vinylic geminal coupling | 0-3 | |
| Vinylic cis coupling | 6-15 | |
| Vinylic trans coupling | 11-18 | |
| Benzylic ortho coupling | 6-10 | |
| Benzylic meta coupling* | 0-4 | |
| Aldehydic proton coupling | 2-3 |
*Actually 4-bond coupling.
Exercise
Predict the splitting patterns of the following molecules:
a)
b)
- Answer
-
a) There are three types of protons, so there will be three peaks. The methyl directly attached to the oxygen will be a singlet. The methylene will be split into a quartet by the neighboring methyl group. The other methyl group will be split into a triplet by the neighboring -CH2-.
b) This molecule is symmetrical, which leads to two types of hydrogens and therefore two peaks. The methylene will be split into a triplet by the two neighboring hydrogens on either side. The -CH- peak will be split into a triplet as well due to the neighboring -CH2-.
How many proton signals would you expect to see in the 1H-NMR spectrum of the structure shown below? For each of the proton signals, predict the splitting pattern. Assume that you see only 3-bond coupling.
- Answer
-
Because of the symmetry in the molecule, there are only four proton signals.
OH proton = singlet
Ha = singlet
Hb = doublet
Hc = doublet
The following spectrum is for C4H10O. Determine the structure. The ratio of integrals from left to right is 2:3.

- Answer
-
The total number of hydrogens is 2+3 = 5. All the hydrogens are not accounted for since we need a total of 10 hydrogens. This means there is symmetry in the molecule. Ten is divisible by five. If we multiply each in the 2:3 ratio, then we get 4:6, which does add to 10.
Typically, integrations of 3 or multiples of 3 are methyl groups. The peak at 1.14 ppm is a methyl group with two neighbors, since the peak is split into a triplet. n +1 = 3, n = 2, n = number of neighbors. The first fragment is a -CH2-CH3. There are two of these fragments, since there are actually 6 total methyl protons.
The peak at 3.43 ppm integrates to 4 and is a quartet. The integration indicates it is a methylene group and since it is a quartet, it must have three neighbors. So far, the fragment would be -CH2-CH3. It is also very far downfield for a proton just bonded to other carbons with hydrogens, which would typically be around 1.2-1.7 ppm (see the table in Section 5.5). This means it must be attached to an electron withdrawing group. The electron withdrawing group in this molecule is the -O-. Our final fragment is CH3-CH2-O-.
If we look at all of our fragments, we can begin to put them together. Let's take the two fragments (-CH2-CH3 and -O-CH2-CH3), each makes one more bond. If you connect them, then you would get CH3-CH2-O-CH2-CH3. The molecule is symmetrical with two types of protons with teh 4:6 (or 2:3) ratio and correct splitting pattern. This spectrum correlates to diethyl ether.
Contributors and Attributions
Dr. Dietmar Kennepohl FCIC (Professor of Chemistry, Athabasca University)
Prof. Steven Farmer (Sonoma State University)
Organic Chemistry With a Biological Emphasis by Tim Soderberg (University of Minnesota, Morris)
Chris P Schaller, Ph.D., (College of Saint Benedict / Saint John's University)

