9.5: Phosphorylation of Alcohols
- Page ID
- 106347
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)A broad family of enzymes called kinases catalyze transfer of a phosphate group from TP to an alcohol acceptor. Mechanistically, the alcohol oxygen acts as a nucleophile, attacking the electrophilic g-phosphorus of TP and expelling ADP.
Glucose is phosphorylated in the first step of the glycolysis pathway by the enzyme hexose kinase (EC 2.7.1.1), forming glucose-6-phosphate.
Hexose kinase mechanism
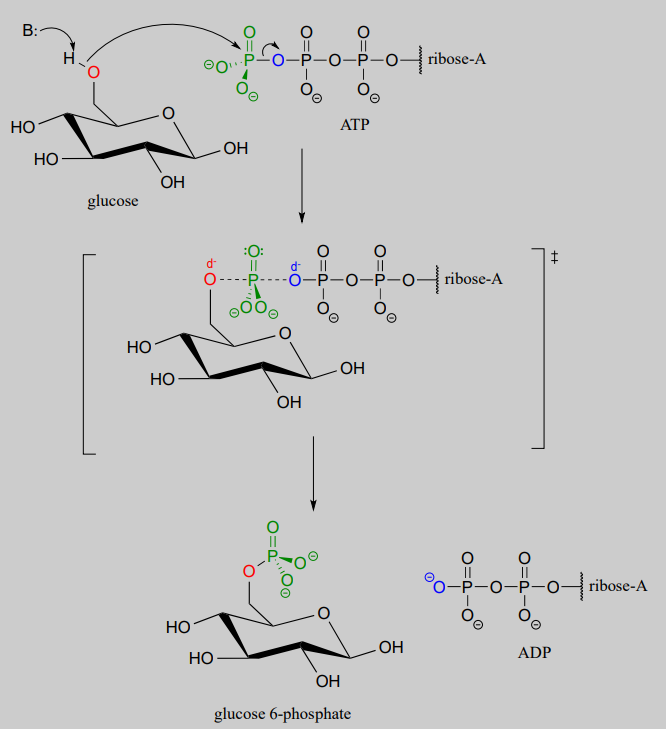
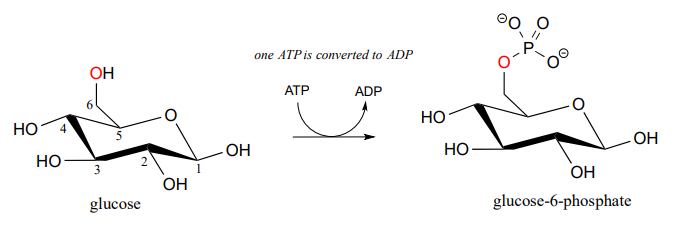
Here is a shorthand way to depict this reaction. Notice the "ATP in, ADP out" notation used below, indicating that ATP is one of the reactants and ADP is one of the products. From here on, we will frequently use this common convention to indicate reaction participants whose structures are not drawn out in a figure.
The biological activity of many proteins is regulated by protein kinases. In a protein kinase reaction, the side chain hydroxyl groups on serine, threonine, or tyrosine residues of certain proteins are phosphorylated by ATP:
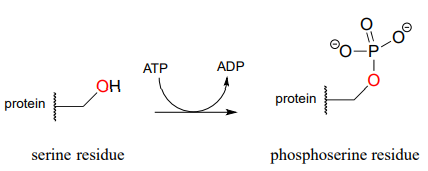
The conversion of a neutral hydroxyl group to a charged phosphate represents a very dramatic change in the local architecture of the protein, potentially altering its folding pattern and ability to bind to small molecules or other proteins. A protein's biological function can be 'switched on' by phosphorylation of a single residue, and switched off again by removal of the phosphate group. The latter reaction we will examine later in this chapter.
- Threonine kinase catalyzes the phosphorylation of the side chain hydroxyl group of threonine residues in proteins. Draw the structure, including the configuration of all stereocenters, of a phosphothreonine residue. Explain how you can predict the stereochemistry of the side chain.
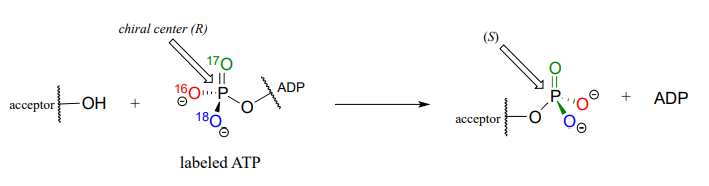
Although stereochemical inversion in phosphate transfer is predicted by theory, the fact that phosphate groups are achiral made it impossible for a long time to verify the phenomenon directly. This was finally accomplished in the late 1970's, when a group of researchers demonstrated phosphate inversion in kinase enzymes using chemically synthesized ATP in which three different isotopes of oxygen were incorporated into the g-phosphate, thus creating a chiral phosphorus center. (Ann. Rev. Biochem. 1980 49, 877).
Alcohols can be converted into organic diphosphates in two different ways. A two-step process simply involves successive transfers of the g-phosphate groups of two ATP donors, such as in these sequential steps in isoprenoid biosynthesis. (EC 2.7.1.36; EC 2.7.4.2). A compound called mevalonate is diphosphorylated in this way in the early phase of the biosynthetic pathway for cholesterol, steroid hormones, and other isoprenoid molecules.
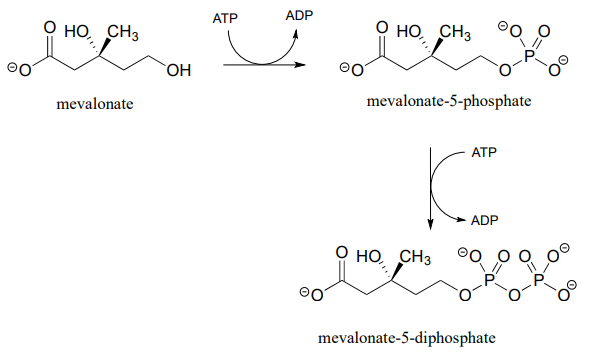
The mechanism for the first phosphorylation step is analogous to that for an alcohol kinase reaction, which we have just seen. In the second phosphate transfer step, catalyzed by a separate enzyme, one of the phosphate oxygens on the organic monophosphate acts as a nucleophilic phosphate acceptor, attacking the g-phosphate of a second ATP.
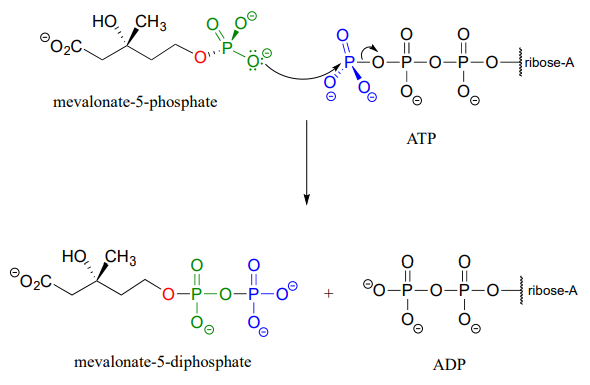
In some metabolic pathways, diphosphorylation occurs by a different mechanism from the one above. stead of sequentially transferring two phosphates from two ATP donors, the alternate mechanism occurs in a single step: the nucleophilic acceptor molecule attacks the b-phosphate of ATP, rather than the g-phosphate. After formation of the trigonal bipyramidal intermediate, it is AMP (not ADP) which is expelled, and what started out as the b and g phosphates of ATP both remain with the acceptor.
In the biosynthesis of DNA and RNA nucleotides, one of the hydroxyl groups on ribose-5-phosphate is diphosphorylated (EC 2.7.6.1) in a one-step mechanism:
A one-step alcohol diphosphorylation reaction (PRPP synthase):
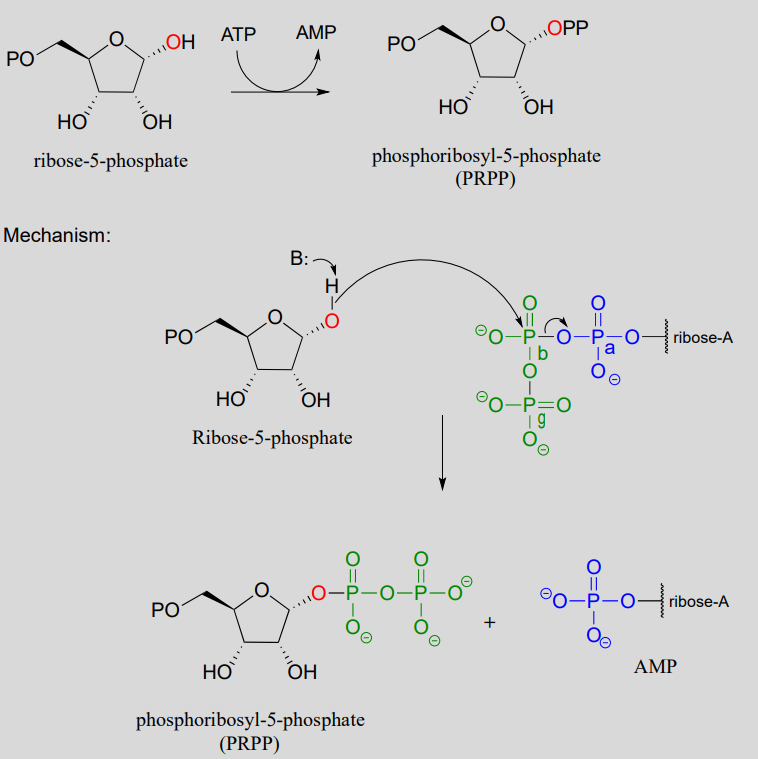
The metabolic role of both of the diphosphorylation processes we just saw is to convert a hydroxyl group into a good leaving group (recall that hydroxide ions are strong bases and poor leaving groups, while phosphates/diphosphates, especially when stabilized in an enzyme active site, are weak bases and very good leaving groups). In nucleoside biosynthesis pathways, the diphosphate group of PRPP acts as a leaving group in the very next metabolic step, which is an \(S_N1\) reaction: we have already seen this reaction in section 8.7).


