25.13: Biosynthesis of Proteins
- Page ID
- 22368
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)One of the most interesting and basic problems connected with the synthesis of proteins in living cells is how the component amino acids are induced to link together in the sequences that are specific for each type of protein. There also is the related problem of how the information as to the amino-acid sequences is perpetuated in each new generation of cells. We now know that the substances responsible for genetic control in plants and animals are present in and originate from the chromosomes of cell nuclei. Chemical analysis of the chromosomes has revealed them to be composed of giant molecules of deoxyribonucleoproteins, which are deoxyribonucleic acids (DNA) bonded to proteins. Since it is known that DNA rather than the protein component of a nucleoprotein contains the genetic information for the biosynthesis of enzymes and other proteins, we shall be interested mainly in DNA and will first discuss its structure. Part or perhaps all of a particular DNA is the chemical equivalent of the Mendelian gene - the unit of inheritance.
The Structure of DNA
The role of DNA in living cells is analogous to that of a punched tape used for controlling the operation of an automatic turret lathe - DNA supplies the information for the development of the cells, including synthesis of the necessary enzymes and such replicas of itself as are required for reproduction by cell division. Obviously, we would not expect the DNA of one kind of organism to be the same as DNA of another kind of organism. It is therefore impossible to be very specific about the structure of DNA without being specific about the organism from which it is derived. Nonetheless, the basic structural features of DNA are the same for many kinds of cells, and we mainly shall be concerned with these basic features in the following discussion.
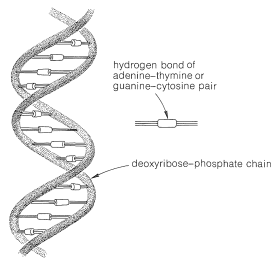
In the first place, DNA molecules are quite large, sufficiently so to permit them to be seen individually in photographs taken with electron microscopes. The molecular weights vary considerably, but values of 1,000,000 to 4,000,000,000 are typical. X-ray diffraction indicates that DNA is made up of two long-chain molecules twisted around each other to form a double-stranded helix about \(20 \: \text{Å}\) in diameter. The arrangement is shown schematically in \(12\):
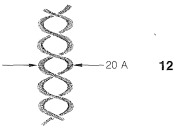
As we shall see, the components of the chains are such that the strands can be held together efficiently by hydrogen bonds. In agreement with this structure, it has been found that, when DNA is heated to about \(80^\text{o}\) under proper conditions, the strands of the helix unwind and dissociate into two randomly coiled fragments. Furthermore, when the dissociated material is allowed to cool slowly under the proper conditions, the fragments recombine and regenerate the helical structure.
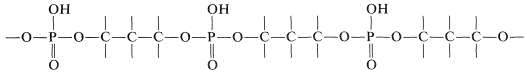
Chemical studies show that the strands of DNA have the structure of a long-chain polymer made of alternating phosphate and sugar residues carrying nitrogen bases, \(13\):

The sugar is \(D\)-2-deoxyribofuranose, \(14\), and each sugar residue is bonded to two phosphate groups by way of ester links involving the 3- and 5-hydroxyl groups:

The backbone of DNA is thus a polyphosphate ester of a 1,3-diol:
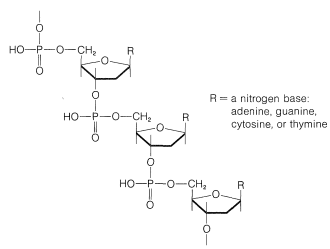
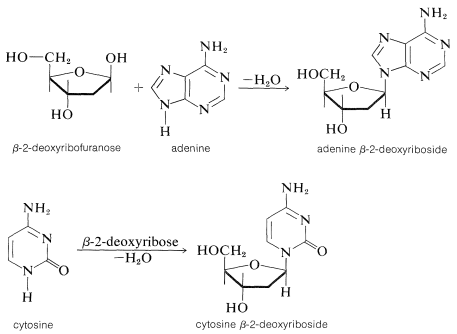
Each of the sugar residues of DNA is bonded at the 1-position of one of four bases: cytosine, \(15\); thymine, \(16\); adenine, \(17\); and guanine, \(18\). The four bases are derivatives of either pyrimidine or purine, both of which are heterocyclic nitrogen bases:
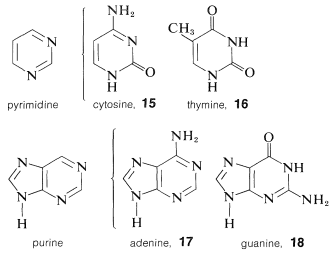
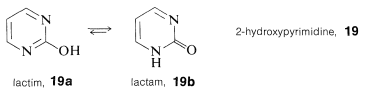
Unlike phenols (Section 26-1), structural analysis of many of the hydroxy-substituted aza-aromatic compounds is complicated by isomerism of the keto-enol type, sometimes called lactim-lactam isomerism. For 2-hydroxypyrimidine, \(19\), these isomers are \(19a\) and \(19b\), and the lactam form is more stable, as also is true for cytosine, \(15\), thymine, \(16\), and the pyrimidine ring of guanine, \(18\).
For the sake of simplicity in illustrating \(\ce{N}\)-glycoside formation in DNA, we shall show the type of bonding involved for the sugar and base components only (i.e., the deoxyribose nucleoside structure). Attachment of 2-deoxyribose is through a \(\ce{NH}\) group to form the \(\beta\)-\(\ce{N}\)-deoxyribofuranoside (Section 20-5):
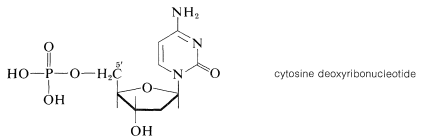
Esterification of the 5'-hydroxyl group of deoxyribose nucleosides, such as cytosine deoxyriboside, with phosphoric acid gives the corresponding nucleotides:\(^{11}\)
The number of nucleotide units in a DNA chain varies from about 3,000 to 10,000,000 Although the sequence of the purine and pyrimidine bases in the chains are not known, there is a striking equivalence between the numbers of certain of the bases regardless of the origin of DNA. Thus the number of adenine (A) groups equals the number of thymine (T) groups, and the number of guanine (G) groups equals the number of cytosine (C) groups: A = T and G = C. The bases of DNA therefore are half purines and half pyrimidines. Furthermore, although the ratios of A to G and T to C are constant for a given species, they vary widely from one species to another.
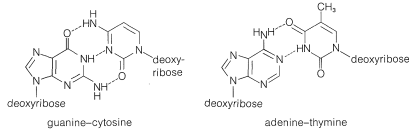
The equivalence between the purine and pyrimidine bases in DNA was accounted for by J. D. Watson and F. Crick (1953) through the suggestion that the two strands are constructed so that, when twisted together in the helical structure, hydrogen bonds are formed involving adenine in one chain and thymine in the other, or cytosine in one chain and guanine in the other. Thus each adenine occurs paired with a thymine and each cytosine with a guanine and the strands are said to have complementary structures. The postulated hydrogen bonds are shown in Figure 25-22, and the relationship of the bases to the strands in Figure 25-23.
Genetic Control and the Replication of DNA
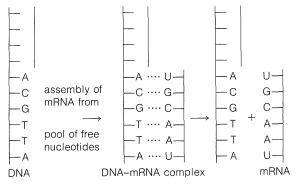
It is now well established that DNA provides the genetic recipe that determines how cells reproduce. In the process of cell division, the DNA itself also is reproduced and thus perpetuates the information necessary to regulate the synthesis of specific enzymes and other proteins of the cell structure. In replicating itself prior to cell division, the DNA double helix evidently separates at least partly into two strands (see Figure 25-24). Each of the separated parts serves as a guide (template) for the assembly of a complementary sequence of nucleotides along its length. Ultimately, two new DNA double strands are formed, each of which contains one strand from the parent DNA.
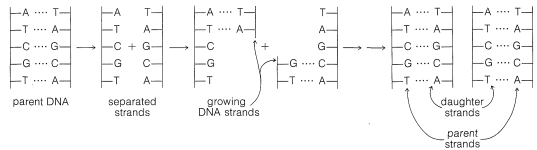
The genetic information inherent in DNA depends on the arrangement of the bases (A, T, G, and C) along the phosphate-carbohydrate backbone - that is, on the arrangement of the four nucleotides specific to DNA. Thus the sequence A-G-C at a particular point conveys a different message than the sequence G-A-C.
It is quite certain that the code involves a particular sequence of three nucleotides for each amino acid. Thus the sequence A-A-A codes for lysine, and U-C-G codes for serine. The sequences or codons for all twenty amino acids are known.
Role of RNA in Synthesis of Proteins
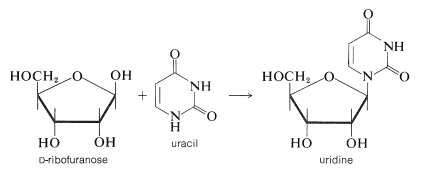
It is clear that DNA does not play a direct role in the synthesis of proteins and enzymes because most of the protein synthesis takes place outside of the cell nucleus in the cellular cytoplasm, which does not contain DNA. Furthermore, it has been shown that protein synthesis can occur in the absence of a cell nucleus or, equally, in the absence of DNA. Therefore the genetic code in DNA must be passed on selectively to other substances that carry information from the nucleus to the sites of protein synthesis in the cytoplasm. These other substances are ribonucleic acids (RNA), which are polymeric molecules similar in structure to DNA, except that \(D\)-2-deoxyribofuranose is replaced by \(D\)-ribofuranose and the base thymine is replace by uracil, as shown in Figure 25-25.
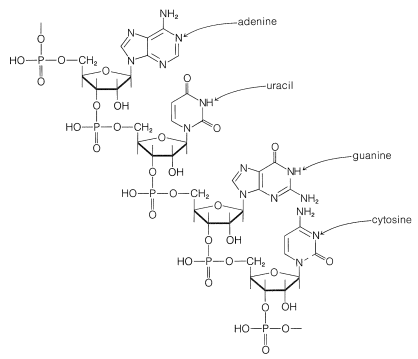
RNA also differs from DNA in that there are not the same regularities in the overall composition of its bases and it usually consists of a single polynucleotide chain. There are different types of RNA, which fulfill different functions. About \(80\%\) of the RNA in a cell is located in the cytoplasm in clusters closely associated with proteins. These ribonucleoprotein particles specifically are called ribosomes, and the ribosomes are the sites of most of the protein synthesis in the cell. In addition to the ribosomal RNA (rRNA), there are ribonucleic acids called messenger RNA (mRNA), which convey instructions as to what protein to make. In addition, there are ribonucleic acids called transfer RNA (tRNA), which actually guide the amino acids into place in protein synthesis. Much is now known about the structure and function of tRNA.
The principal structural features of tRNA molecules are shown schematically in Figure 25-26. Some of the important characteristics of tRNA molecules are summarized as follows.
1. There is at least one particular tRNA for each amino acid.
2. The tRNA molecules have single chains with 73-93 ribonucleotides. Most of the tRNA bases are adenine (A), cytosine (C), guanine (G), and uracil (U). There also are a number of unusual bases that are methylated derivatives of A, C, G, and U.
3. The clover-leaf pattern of Figure 25-26 shows the general structure of tRNA. There are regions of the chain where the bases are complementary to one another, which causes it to fold into two double-helical regions. The chain has three bends or loops separating the helical regions.
4. The 5'-terminal residue usually is a guanine nucleotide; it is phosphorylated at the 5'-\(\ce{OH}\). The terminus at the 3' end has the same sequence of three nucleotides in all tRNA's, namely, CCA. The 3'-\(\ce{OH}\) of the adenosine in this grouping is the point of attachment of the tRNA to its specific amino acid:
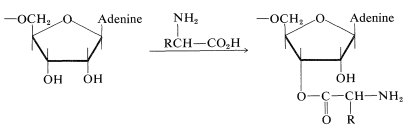 \(\tag{25-9}\)
\(\tag{25-9}\)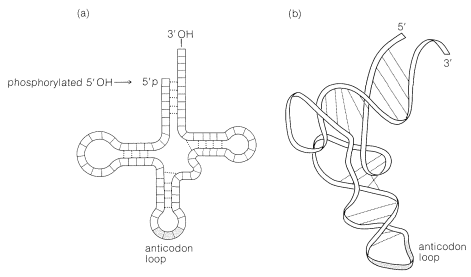
With this information on the structure of tRNA, we can proceed to a discussion of the essential features of biochemical protein synthesis.
The information that determines amino-acid sequence in a protein to be synthesized is contained in the DNA of a cell nucleus as a particular sequence of nucleotides derived from adenine, guanine, thymine, and cytosine. For each particular amino acid there is a sequence of three nucleotides called a codon.
The information on protein structure is transmitted from the DNA in the cell nucleus to the cytoplasm where the protein is assembled by messenger RNA. This messenger RNA, or at least part of it, is assembled in the nucleus with a base sequence that is complementary to the base sequence in the parent DNA. The assembly mechanism is similar to DNA replication except that thymine (T) is replaced by uracil (U). The uracil is complementary to adenine in the DNA chain (see Figure 25-27). After the mRNA is assembled, it is transported to the cytoplasm where it becomes attached to the ribosomes.
The amino acids in the cytoplasm will not form polypeptides unless activated by ester formation with appropriate tRNA molecules. The ester linkages are through the 3'-\(\ce{OH}\) of the terminal adenosine nucleotide (Equation 25-9) and are formed only under the influence of a synthetase enzyme that is specific for the particular amino acid. The energy for ester formation comes from ATP hydrolysis (Sections 15-5F and 20-10). The product is called an aminoacyl-tRNA.
The aminoacyl-tRNA's form polypeptide chains in the order specified by codons of the mRNA bound to the ribosomes (see Figure 25-28). The order of incorporation of the amino acids depends on the recognition of a codon in mRNA by the corresponding anticodon in tRNA by a complementary base-pairing of the type A \(\cdots\) U and C \(\cdots\) G. The first two bases of the codon recognize only their complementary bases in the anticodon, but there is some flexibility in the identity of the third base. Thus phenylalanine tRNA has the anticodon A-A-G and responds to the codons U-U-C and U-U-U, but not U-U-A or U-U-G:
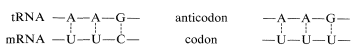
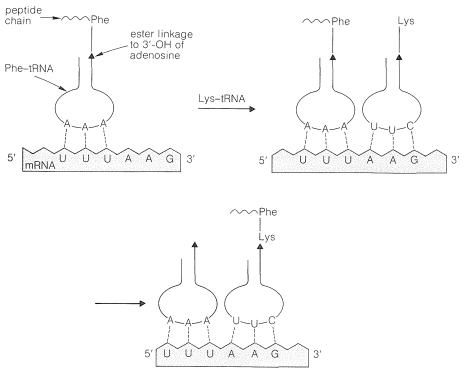
The codons of the mRNA on the ribosomes are read from the 5' to the 3' end. Thus the synthetic polynucleotide (5')A-A-A-(A-A-A)\(_n\)-A-A-C(3') contains the code for lysine (A-A-A) and asparagine (A-A-C); the actual polypeptide obtained using this mRNA in a cell-free system was Lys-(Lys)\(_n\)-Asn, and not Asn-(Lys)\(_n\)-Lys.
The start of protein synthesis is signaled by specific codon-anticodon interactions. Termination is also signaled by a codon in the mRNA, although the stop signal is not recognized by tRNA, but by proteins that then trigger the hydrolysis of the completed polypeptide chain from the tRNA. Just how the secondary and tertiary structures of the proteins are achieved is not yet clear, but certainly the mechanism of protein synthesis, which we have outlined here, requires little modification to account for preferential formation of particular conformations.
\(^{11}\)The positions on the sugar ring are primed to differentiate them from the positions of the nitrogen base.
Contributors and Attributions
John D. Robert and Marjorie C. Caserio (1977) Basic Principles of Organic Chemistry, second edition. W. A. Benjamin, Inc. , Menlo Park, CA. ISBN 0-8053-8329-8. This content is copyrighted under the following conditions, "You are granted permission for individual, educational, research and non-commercial reproduction, distribution, display and performance of this work in any format."


