14: Radical Detection17
- Page ID
- 150534
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Name: ______________________________
Section: _____________________________
Student ID#:__________________________
Template:HideTOCDetection of Radicals
Unpaired electrons can provide a diagnostic tool in the study of many systems. The interaction of unpaired electrons with magnetic fields is useful.
Two Important tools: 1) Magnetic moments and 2) EPR spectroscopy.
Review: Measurement of Magnetic Moments
- Compounds that have unpaired electrons are said to be ___________ (paramagnetic / diamagnetic ).
- Compounds that have no unpaired electrons are said to be be ___________ (paramagnetic / diamagnetic ).
- ___________ (paramagnetic / diamagnetic ) compounds are attracted to a magnetic field.
In a Gouy balance, a sample is suspended from a balance such that the sample tube is half in and half out of the field of a very strong electromagnet located below the balance. First, the sample is weighed with the electromagnet turned off. Then, the electromagnet is turned on and the sample is reweighed.
- If the sample is truly paramagnetic, the sample will be _______ (attracted/repelled) by the magnetic field and as a result the weight of the sample will ___________(increase/decrease) when the magnetic field is applied. This quantity is called the gram susceptibility.
The corrected molar magnetic susceptibility, XM, can be obtained which is converted to the effective magnetic moment using the following equation:
$$
\mu_{eff}=2.83 \sqrt{X_{M} T} \quad \mathrm{T}=\text { temperature in Kelvin. }
\nonumber \]
This number is compared to a predicted value, known as the spin–only magnetic moment, μso.
$$
\mu_{\infty}=\sqrt{n(n+2)} \quad n=\text { number of unpaired electrons. }
\nonumber\]
For most first row transition metal complexes there should be good match between μso and μeff.
$$
\text { Number of unpaired electrons } \approx \mu_{\mathrm{so}}-1
\nonumber\]
Practice using magnetic moment measurements:
- To determine whether a complex is high spin or low spin:
- Draw the splitting diagram both a low spin and a high spin octahedral Fe(II) complex.
- Predict μso for each of these complexes.
- To determine the oxidation state of a metal. Some metals have several common oxidation states, or are easily oxidized such that the oxidation state of a metal may change under the reaction conditions.
- Predict μso for Fe(III) high spin and low spin (show splitting diagrams).
- How do these values compare to the Fe(II) values?
- Could you determine if the metal is Fe(II) or Fe(III)?
Electron Paramagnetic Resonance (epr)
Unpaired electrons have a useful feature: spin. Like protons and certain other nuclei, this spin can be detected via its interaction with a magnetic field.
Spin (on either an electron or a proton) has two possible values: up and down.
- Fill in the spin: _____up _____down
We probably have lots of radicals in one sample, rather than one molecule, so we will deal with a population of them. Ordinarily, up and down spins have the same energy.
- Fill in the spins (five of each):
In a sample, the spins are random.
In an epr experiment, the sample is placed in a magnetic field. In the external field, the spins tend to either align with the magnetic field or line up against the magnetic field.
- Add several spins into the sample tube.
In a magnetic field, the two spin states have slightly different energies. A narrow majority of electrons will adopt the lower energy state.
- Fill in the spins in the presence of a magnetic field:
Microwave-frequency radiation is sent into the sample, and a few of the electrons are promoted to the next level.
- Fill in the spins after irradiation:
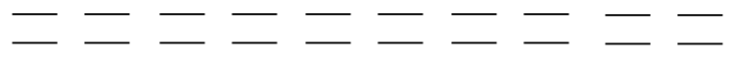
The splitting between spin states depends on the strength of the magnetic field experienced by the electron. In the cartoon, the length of the photon represents its energy.
- Circle the point in the cartoon at which the radiation will be absorbed.
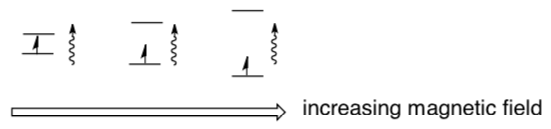
EPR measures wavelength of photon absorbed by these electrons when they are promoted. The frequency is mostly dependent on the atom on which the unpaired electron is found.
EPR spectra can be generated by either varying the photon frequency incident on a sample while holding the magnetic field constant or doing the reverse. In practice, it is usually the frequency that is kept fixed.
- The x-axis in an epr spectrum is called the g-factor. It is a measure of the __________ ( magnetic field strength / intensity ).
- The y-axis in an epr spectrum is called the signal. It is a measure of the __________ ( magnetic field strength / intensity ).
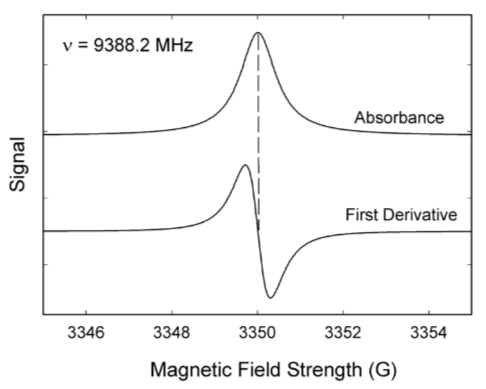
The local field experienced by the electron is influenced by spins of nearby nuclei. Carbon (12C) does not have an overall spin, but other nuclei do.
To calculate the effect of the nuclear spin of the atom next door, use the following formula:
Multiplicity = 2 n I +1
where n = the number of nuclei
I = nuclear spin
- Suppose a radical is centered on a carbon with one hydrogen ( I= 1⁄2 ) attached (one spin, up or down). Calculate the multiplicity using the formula.
- How many different magnetic fields can the electron experience?
- The following simulated spectrum is for a CH3 radical. Explain the number of peaks, and the ratio in which they occur.
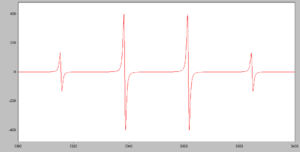
(http://en.Wikipedia.org/wiki/Electro...etic_resonance)
NOTE: The peaks in epr spectra have peculiar “up & down” shapes.
- Sketch expected epr spectra for the following radicals (looking at the number of hydrogens on the neighboring atoms):
- •CH2OH (16O has no spin)
- (C6H5)3C•
Radicals may also be affected by more distant nuclei, although the effect is smaller with distance.
- The following simulated spectrum is for a CH2(OCH3) radical. Explain the pattern of peaks.
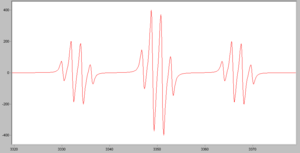
The following ions have an unpaired electron. In each case, the nuclear spin of the major isotope is provided.
- Sketch expected epr spectra for each case.
- Cu(II) (3/2)
- Fe(III) (0)
- V(IV) (7/2)
In a more complicated case, Mo has 75% abundance of isotopes with spin = 0 and 25% abundance of isotopes with spin = 5/2.
- Sketch the epr spectrum of a radical on Mo(V).
Integrative problem: Cytochrome P-450
Cytochrome P-450 enzymes are responsible for the initial steps in the metabolism of 75% of known pharmaceuticals when used in the human body. In general, it acts by using molecular oxygen to hydroxylate C-H bonds, as shown in the generic reaction below. Though it is one of the most studied molecules in all of chemistry and biochemistry, the precise way in which it performs the reaction remains a mystery!
R-H + 0.5 O2 -> ROH
The basic structure of the active site of the enzyme is shown below. On the left is shown a planar array of ligands around the iron called a porphyrin. It is similar to the environment around the iron in hemoglobin. There is a fifth ligand, a sulfide residue from an amino acid, that connects the iron-porphyrin complex to the enzyme. This is shown in the side view of the complex on the right.
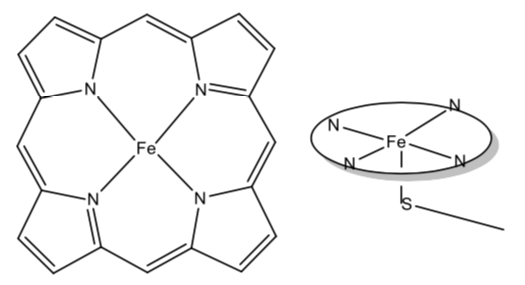
- Initially, the Fe is in the +3 oxidation state. Show how the formal charges on the ligands are consistent with this assignment.
- Draw the d orbital splitting diagram for the complex. If μ is 1.7, how should the electrons be placed in the diagram?
- Cytochrome P-450 gets its name from the fact that its carbon monoxide complex absorbs strongly at 450 nm. What color would the complex be?
- The complex (without CO) absorbs at about 420 nm and has e ~ 50,000 M-1 cm-1. What sort of electronic transition is the likely source of this absorbance? If you are not sure, discuss the possibilities.
When this enzyme reacts with O2 or other oxygen atom donors, a complex is formed that has been suggested to be that shown below. It is proposed that this is an Fe(IV) complex, with the porphyrin also oxidized by one electron. Thus there is an additional unpaired electron associated with the porphyrin.
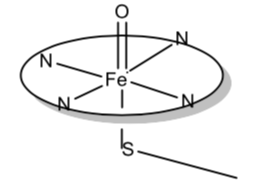
- Why is the porphyrin ligand able to stabilize an unpaired electron?
- Draw the d orbital splitting diagram for Fe(IV) and place the proper number of electrons in the diagram.
- What should the total magnetic susceptibility of the complex be?
The absorbance spectrum of this complex is dominated by an intense peak at approximately 360 nm.
- Compare this to the original peak at 420 nm.
- Is a shift in this direction consistent with going from 5 to 6 ligands?
- Is a shift in this direction consistent with going from Fe(III) to Fe(IV)?
It is believed that in addition to tethering the metal complex to the rest of the enzyme, the role of the sulfur is to stabilize Fe in this high oxidation state.
- How/why is sulfur able to do this??
It has been proposed that the oxygen on the Fe(IV) complex inserts itself into a C-H bond of a hydrocarbon chain in a pharmaceutical to make a COH (hydroxyl) group. This reaction aids in removal of the molecule from the body. How?
Application Problem—Spin Crossover Complexes
Thus far, we have tended to think of transition metal complexes as being either high spin or low spin. However, there are complexes that exist as a mixture of high spin and low spin complexes, or a complex that exists as a high spin complex at one temperature and a low spin complex at a different temperature.
Klaui, et. al, Inorganic Chemistry, 1987, 25, 3977-3982.
- Draw the electron arrangement for high spin and low spin Co(III). Predict the value of μeff in either case.
- Co(III) complexes are virtually always low spin. The only common high spin Co(III) complexes are [Co(F)6]3- and [Co(F)3(H2O)3]. Use your knowledge of the spectrochemical series to predict the formulas for two other complexes (all six ligands identical) that should be high spin.
- In fact the complexes you predicted to be high spin do not exist. This is because Co(III) is capable of oxidizing those ligands to X2. Explain why F- should be more inert to oxidation.
The authors prepared a series of complexes as shown below. (Although they are metal complexes themselves, they serve as ligands for the Co (III) ions that are being studied.) Note that overall, the ligand is an anion.
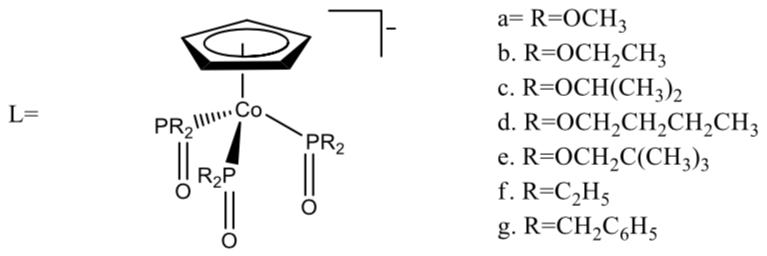
Two of these ligands bond to Co(III) through the oxygens to form an octahedral complex of the general formula [CoL2]+.
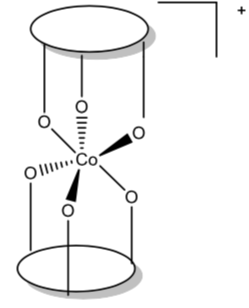
The ligand field strength of L depends on steric and electronic factors.
- As R gets larger, what will happen to the O-Co bond, and how will this affect the strength of L as a ligand? What will this tend to do to the spin state of the resulting complex?
- As R becomes a better electron donor, what will happen to the strength of L as a ligand? What will this tend to do to the spin state of the resulting complex?
The authors measured the magnetism of the Co(III) complexes with ligands 1f and 1g. The magnetic moment was approximately 0 and was independent of temperature over the range 10-320 K. On the other hand the compounds with R=O-alkyl were paramagnetic and green and room temperature but become yellow and diamagnetic as the temperature is lowered. The change in magnetism and color is completely reversible.
- According to the authors “the paramagnetic behavior dominates in [the complexes of 1c] and decreases in the order 1c>1e>1d>1b>1a.” Of this series, which is the weakest ligand? The strongest? Which of the factors (sterics or electronics) provide the best explanation for the ligand field strengths of L?
- The complexes where L=1a-1e are thus low spin at low temperature and high spin at some higher temperature. Why do you suppose it is this way rather than the other way around? (As the temperature is raised, what happens to the Co-L bond, and why would this tend to make Δ smaller? Hint—think IR spectroscopy.)
In order to further understand the complexes, the authors gathered uv-vis spectra for the complexes. In this case, they were studying the Co(II) complexes rather than Co(III).
- One peak was measured at ~340 nm and had e= 3000 M-1 cm-1. What sort of transition causes this?
- The Co(II) complexes had three additional peaks. The energy of the lowest energy peak is given in the table below. Is this consistent with the ranking of the ligands based on magnetic measurements as discussed previously?
L ν (cm-1) 1a 7150 1b 7200 1c 7050 1d 7170 1f 7200


