13: Nitrogen Fixation
- Page ID
- 150531
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Name: ______________________________
Section: _____________________________
Student ID#:__________________________
Nitrogen Fixation
Passport
Nitrogen fixation is a process by which nitrogen (N2) in the atmosphere is converted into ammonia (NH3).
- Draw a Lewis Structure for N2.
- Will the conversion of N2 -> NH3 be an oxidation or reduction? Show the oxidation state of the nitrogen atoms in the starting materials and products.
- Write a balanced equation for process of converting N2 -> NH3.
Nitrogen fixation, natural and synthetic, is essential for all forms of life because nitrogen is required to biosynthesize basic building blocks of life such as DNA and proteins.
Therefore nitrogen fixation is essential for agriculture and the manufacture of fertilizer.
Extra Credit Opportunity: (instructor dependent)
- Listen to: Radiolab podcast on Fritz Haber
- Submit a summary of the chemistry developed by Fritz Haber. What were the implications of the development of this new catalytic cycle? Describe the other activities in which he was involved and some ethical issues.
Other sources to supplement this podcast:
Nobel Website Biography
Chemical Heritage Foundation
V. Smil, Nature, July 29, 1999, p 415
Synthetic Nitrogen Fixation: Haber-Bosch Process
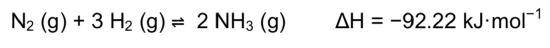
Nitrogen (N2) is very unreactive.
- Suggest a reason why.
- Write an equation for the Keq for the Haber-Bosch Process.
Keq =
Table 1. Variation in Keq for the Haber-Bosch Process as a function of temperature.
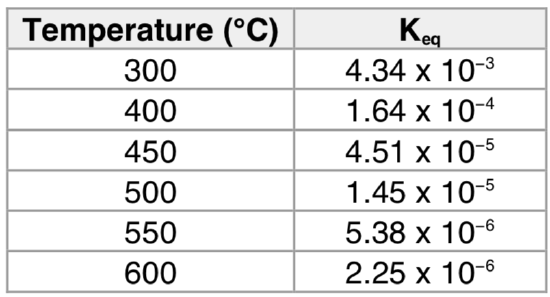
- What happens to the Keq with increasing temperature?
- Why? What factor is the causing the shift in Keq?
Based on your understanding of thermodynamics and kinetics:
- Is this reaction thermodynamically favored?
- Is it kinetically favored? (ie How high is the Ea?)
- Draw a potential energy diagram for this process.
- Using Le Chatelier's principle, how could you drive this reaction toward products?
Mechanism of the Haber-Bosch Process
The Haber process relies on catalysts that accelerate the scission of this triple bond.
The Haber-Bosch Process allows ammonia to be synthesized from nitrogen (in air) and hydrogen (usually made via steam reforming of natural gas). The use of the resulting ammonium salts as fertilizers has helped to feed people all over the world.
A simplified, proposed mechanism for the Haber-Bosch Process is given below. The reaction takes place on the surface of mixed metal/metal oxides. Usually iron is used, although ruthenium and osmium have also been employed.
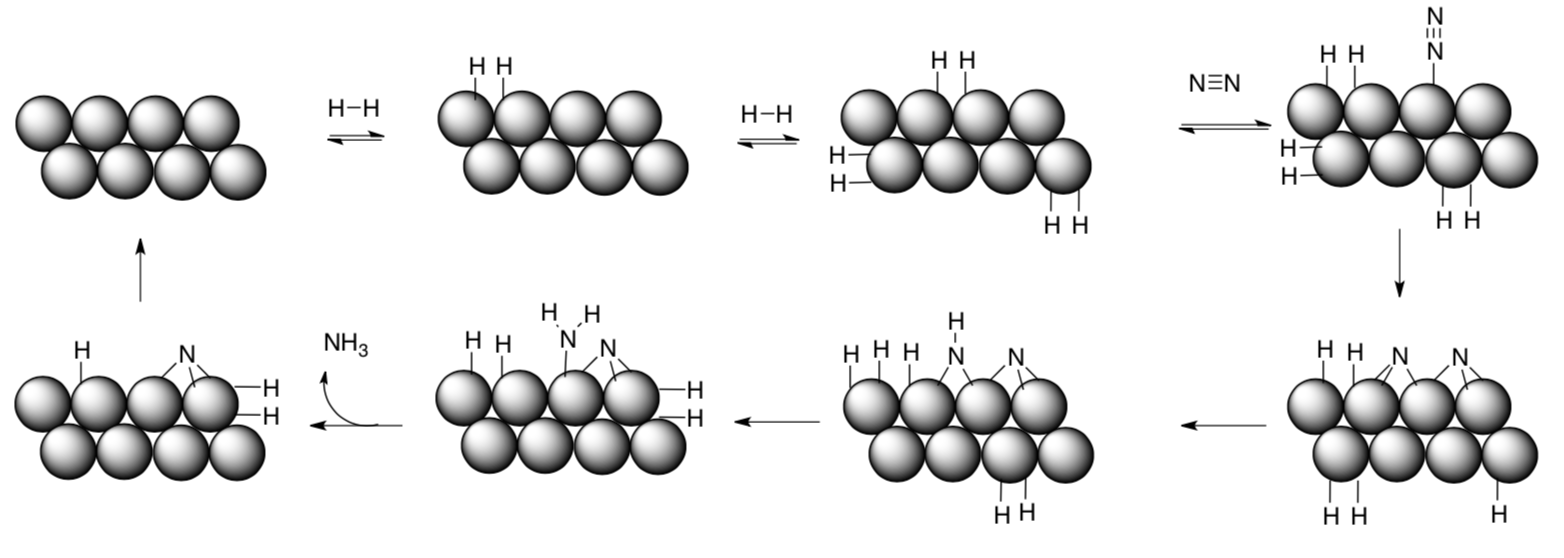
- Is this (heterogeneous/homogeneous) catalysis. Circle one.
- Label steps in the above scheme (association, oxidative addition, etc).
- Overall, the process represents reduction / oxidation of N2 (circle one).
- Why is it not surprising that iron, ruthenium and osmium might all catalyze the same reaction?
- What is the practical advantage of using iron rather than ruthenium or osmium?
Nitrogen Fixation in Organisms
Passport
Nitrogen plays a key role in many biological structures.
- Which structural types CAN contain nitrogen?
Sugars | Proteins | Lipids | Nucleic Acids
- Which structural types ALWAYS contain nitrogen?
Sugars | Proteins | Lipids | Nucleic Acids
- Draw an amino acid.
- In these structures, the nitrogen atoms are typically bound to carbon and hydrogen. It is also usually in a (oxidized OR reduced) form.
- Yet in the environment, nitrogen is usually inorganic (N2, NO3-). It is usually in (oxidized OR reduced) form.
Nitrogen uptake from the environment proceeds through two pathways – nitrate assimilation and nitrogen fixation.
- Balance the reactions involved in these two pathways:
Nitrate Assimilation is a two-step metabolic pathway that reduces NO3- used by various plants, fungi and bacteria. This accounts for 99% of the nitrogen uptake by organisms.
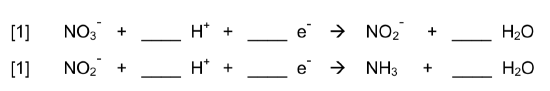
Nitrogen Fixation is the reduction of nitrogen gas via an enzyme found only in prokaryotic cells.

- How do large animals get their nitrogen?
Nitrogen Cycle
Extra Credit
The nitrogen cycle is the process by which nitrogen is converted between its various chemical forms.
Nitrogen Cycle:
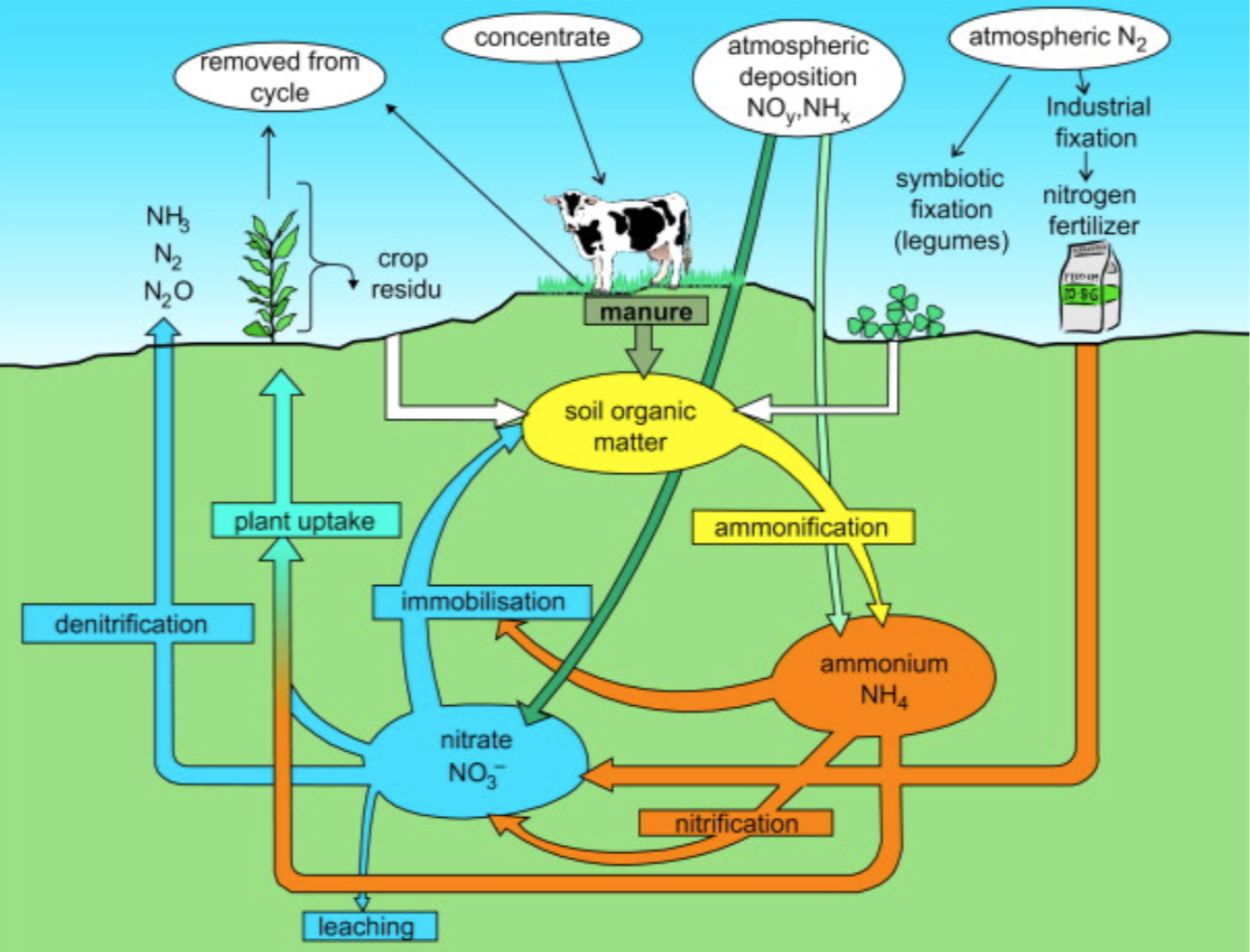
from: J.W. Erisman, A. Bleeker, J. Galloway, M.S. Sutton, Environmental Pollution,2007, 150(1), 140-149.
Imbalance of the Nitrogen Cycle
The nitrogen cycle is of particular interest because nitrogen availability can affect the rate of key ecosystem processes, including decomposition. As a result of extensive cultivation of legumes (particularly soy, alfalfa, and clover), growing use of the Haber- Bosch process in the creation of chemical fertilizers, and pollution emitted by vehicles and industrial plants, human beings have more than doubled the annual transfer of nitrogen into biologically-available forms.
- Report on one of the following impacts of an altered nitrogen balance in the ecosystem (be sure to include at least one chemical equation):
- Agricultural productivity
- Nutrient cycling and ammonification
- Plant species diversity
- Freshwater acidification
- Eutrophication of marine systems
Biological Nitrogen Fixation
80% of the majority the atmosphere is nitrogen. However, atmospheric nitrogen has limited availability for biological use, leading to a scarcity of usable nitrogen in many types of ecosystems.
Microorganisms that fix nitrogen are bacteria called diazotrophs. Some higher plants, and some animals (termites), have formed associations with diazotrophs.
The advantage to the plants is that they have access to a nitrogen source.
- What might be the advantage to the symbiont?
Biological nitrogen fixation occurs when atmospheric nitrogen is converted to ammonia by an enzyme called nitrogenase. This process is similar to the Haber-Bosch process.
- Balance the reaction performed by nitrogenase:
N2 +8H+ +_______e- → 2NH3 +_____H2
- How does this equation differ from the Haber-Bosch process?
The process is coupled to the hydrolysis of 16 equivalents of ATP and is accompanied by the co-formation of one molecule of H2.
- Based on Haber-Bosch, is this reaction endothermic OR exothermic?
- Why is ATP needed?
Biological Nitrogen Fixation:
- Draw a series of likely intermediates for dinitrogen reduction to two ammonia molecules, naming each intermediate.
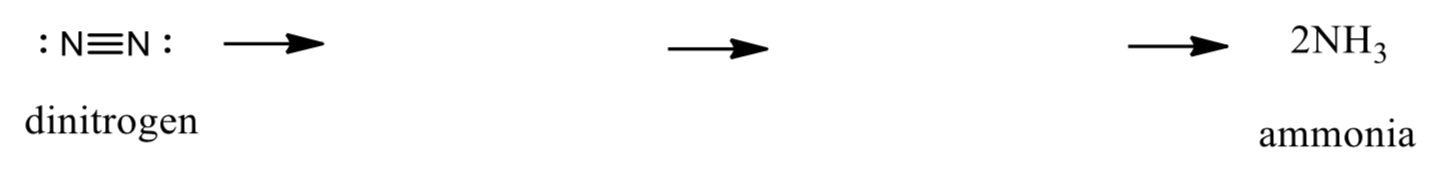
- What is the bond order in each? Which step is most difficult?
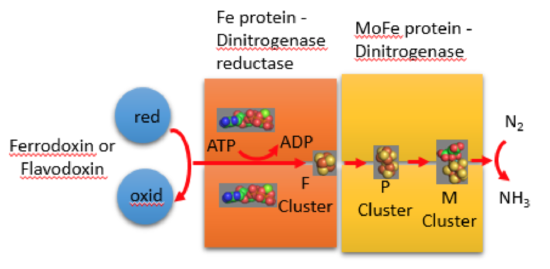
Diazotrophs use nitrogenase enzymes to fix atmospheric nitrogen gas.
Nitrogenase consists of the following proteins/cofactors: the Fe protein, which contains a [Fe4S4] cluster, and the MoFe protein, which contains two different classes of metal
cluster: P-cluster ([Fe8S7]) and FeMo cofactor.
Biological Nitrogen Fixation: MoFe Cofactor
Nitrogenase contains a FeMo cofactor that is still poorly characterized and the understood.
- Calculate the oxidation state of the metal centers in FeMo Cofactor shown below.
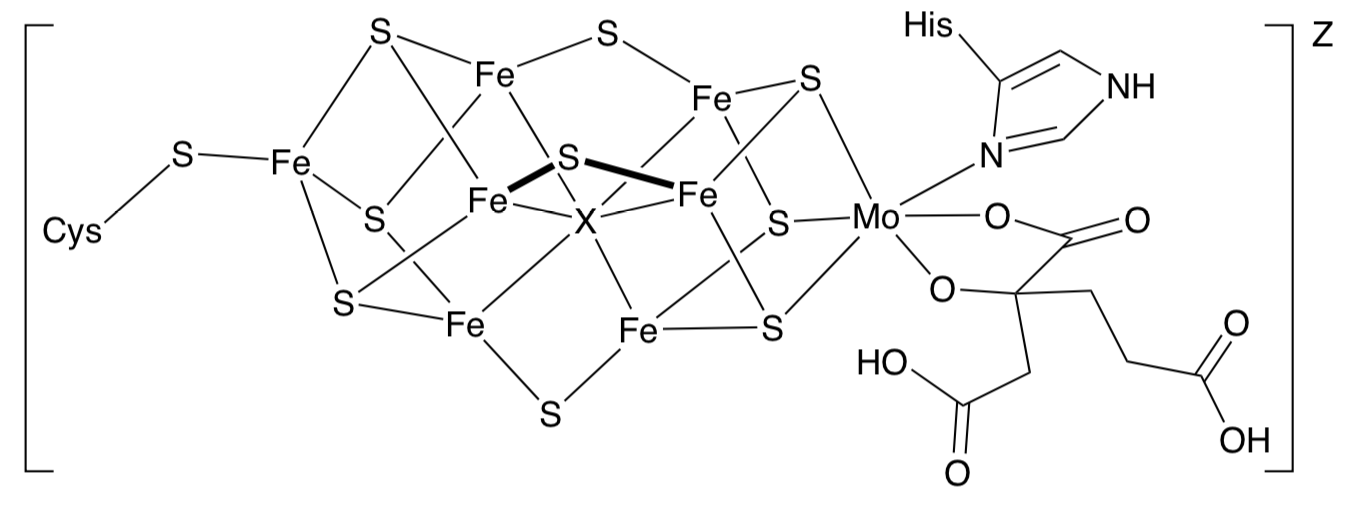
from Xu-Dong Chen, Wei Zhang, Jeremiah S. Duncan, and Sonny C. Lee,Inorganic Chemistry, 2012, 51, 12891-12904.
- Assume X is monoatomic C. What would the charge on that ligand be?
- Assume the His is neutral. What is the total charge on ligands?
- How many Fe atoms are involved?
The charge state, z, of the entire cluster is unknown but the resting state of the cofactor has an odd electron count.
- If z = -3, what must be the charge on all of the metal atoms?
- The Mo is known to be +4. What is the overall charge to be distributed on the Fe atoms?
- What is the charge on a typical Fe atom?
- What is the geometry of each Fe atom?
- The sulfur ligands are
Weak field Or Medium field Or Strong field
- Would these be low spin or high spin Fe centers?
- Show a LFT splitting diagram for one of these Fe centers.
Overall Nitrogenase Scheme
The Lowe and Thorneley (LT) proposed reaction scheme is shown below.
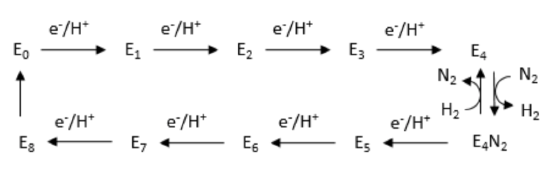
Nitrogen is binding to one of the Fe centers in the FeMo cofactor.
- What type of donor is N2 (circle one)?
Sigma only | Pi donor | Pi acceptor
- Starting from E0, why are so many electrons added before the N2 can easily bind?
- Why can many electrons be accepted by the FeMo cofactor?
Biological Nitrogen Fixation: Catalytic Cycle
Blöchl has proposed a catalytic cycle for the nitrogenase mechanism in biological nitrogen fixation based on extensive computational modeling.
From Blöchl, et.al., http://www2.pt.tu-clausthal.de/atp/p.../nitro.htmlAnd Kästner and Blöchl, J. Am. Chem. Soc., 2007, 129, 2998-3006.
- Add arrows for electron transfer. Use single-headed arrows for single electron movements.

- Fill in the missing metal complexes.
- Add oxidation states to the metal centers.
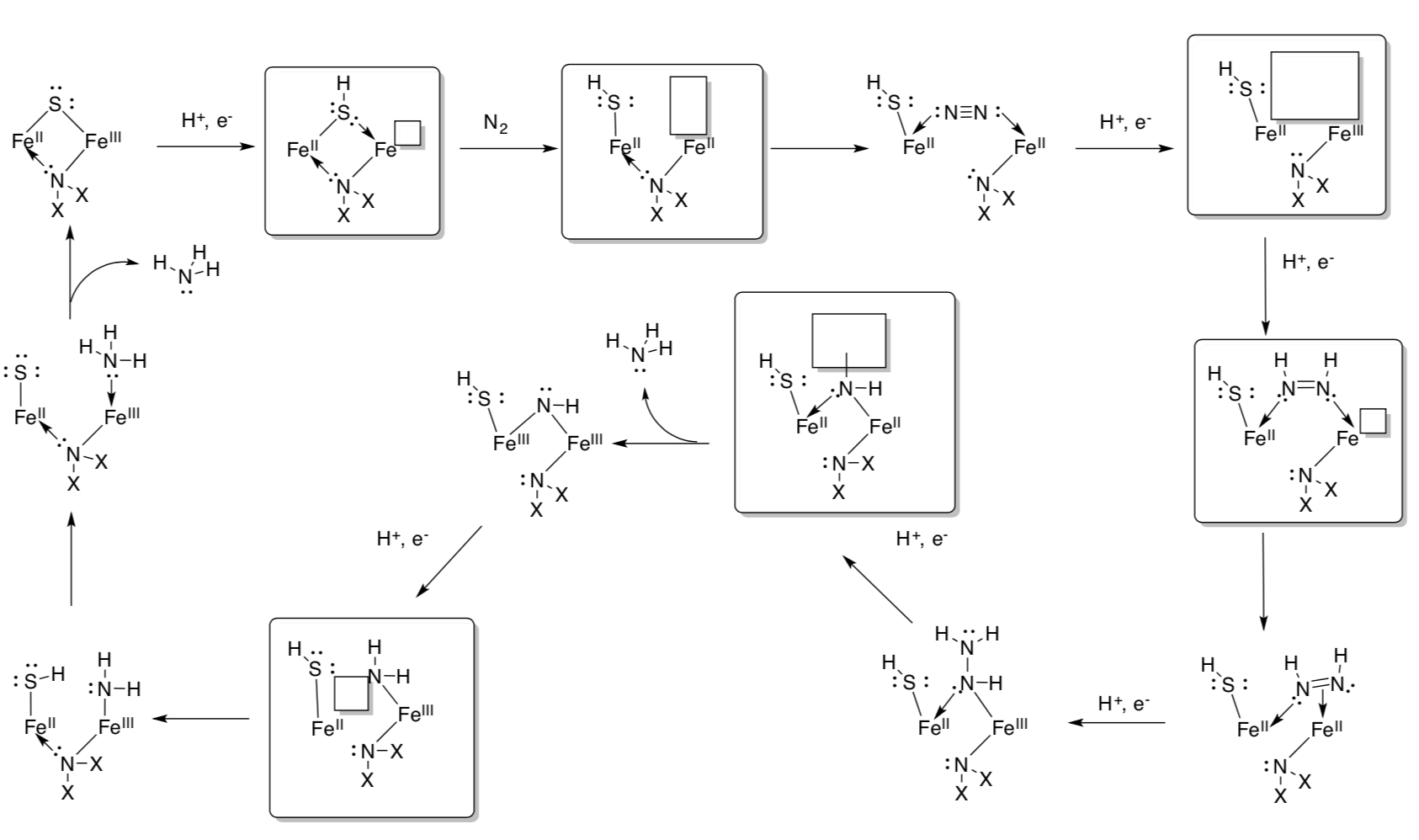
New Developments in Mechanistic Work on Nitrogenase
Overall, the reduction of dinitrogen to ammonia is a six-electron, six-proton process.
- Fill in the intermediate reduction states (diazene and hydrazine).
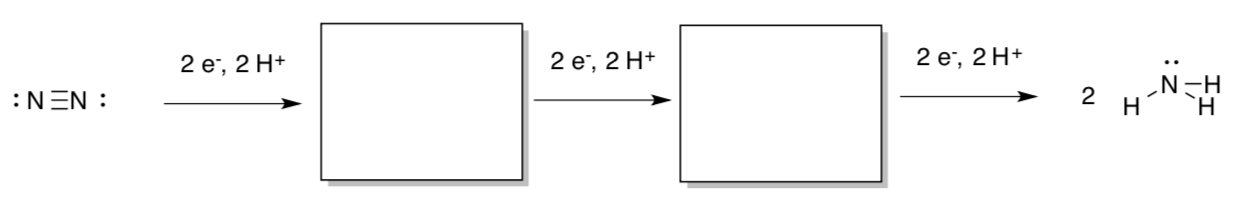
However, the overall reaction has long been known to involve additional protons.
N2 + 8H+ + 8e- -> 2 NH3 + H2
Recent work, notably by inorganic spectroscopist Brian Hoffman (Northwestern), indicates where the H2 comes from. New evidence from ENDOR studies (similar to NMR and EPR) suggests iron hydrides are present in nitrogenase.
An iron hydride can form via protonation of iron.
- Show the change in oxidation state that results.
- Provide a name for this organometallic mechanism type.
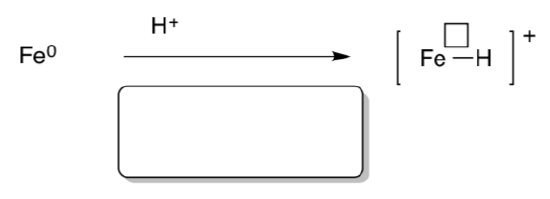
Dihydrogen may result from iron hydrides, below.
- Show the changes in oxidation state that result.
- Provide names for these organometallic mechanism types.
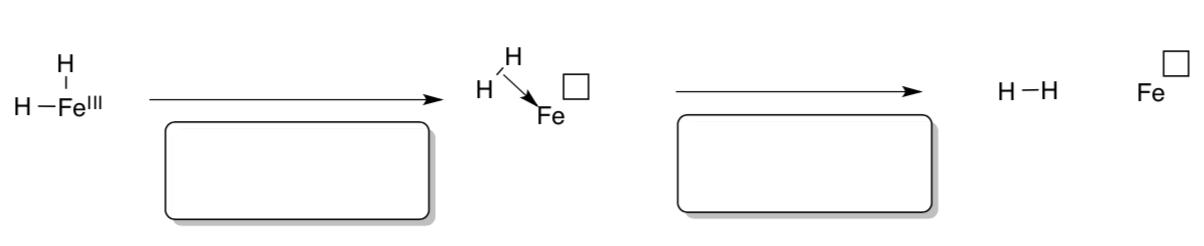
There is evidence that reactions like this one occur via “sigma-complex” intermediates (shown, middle). In this complex, electrons in the s-orbital of H2 are donated to the metal.
The drawing below shows the FeMo cofactor and its surroundings. The nitrogen is believed to enter the protein from the top of this picture.
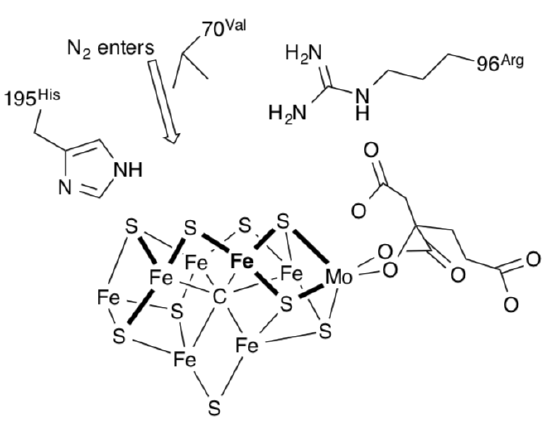
- Site directed mutagenesis shows that dinitrogen uptake stops if the Val (R = CHMe2) is replaced with Ile (R = CHMeCH2Me). Suggest a reason why.
- Site directed mutagenesis shows no reaction occurs if the His is replaced. Suggest a role for the histidine, based on the equation for the reaction.
The catalytic reaction is thought to occur along the top face of the cofactor, isolated in the simpler picture below.
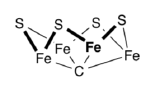
- Why is it reasonable to assume the reaction happens in this part of the cofactor?
Additional evidence leads researchers to think dinitrogen binds at the iron in bold.
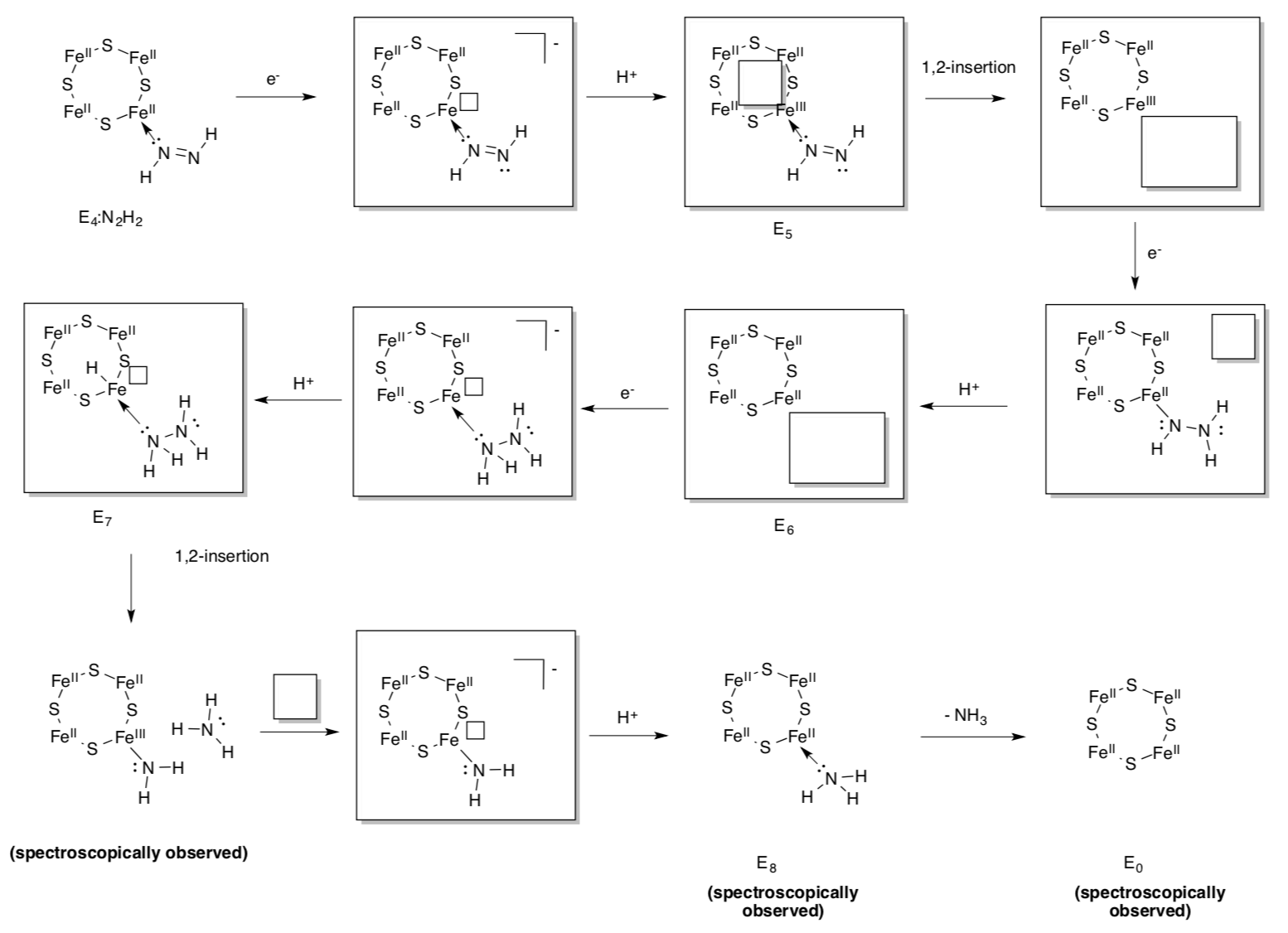
Spectroscopic data suggest the cofactor proceeds through a number of redox/protonation states during the catalytic cycle. These states are called E0, E1,...E8.
- The first four states are shown below. Fill in the missing pieces (signaled by the boxes).
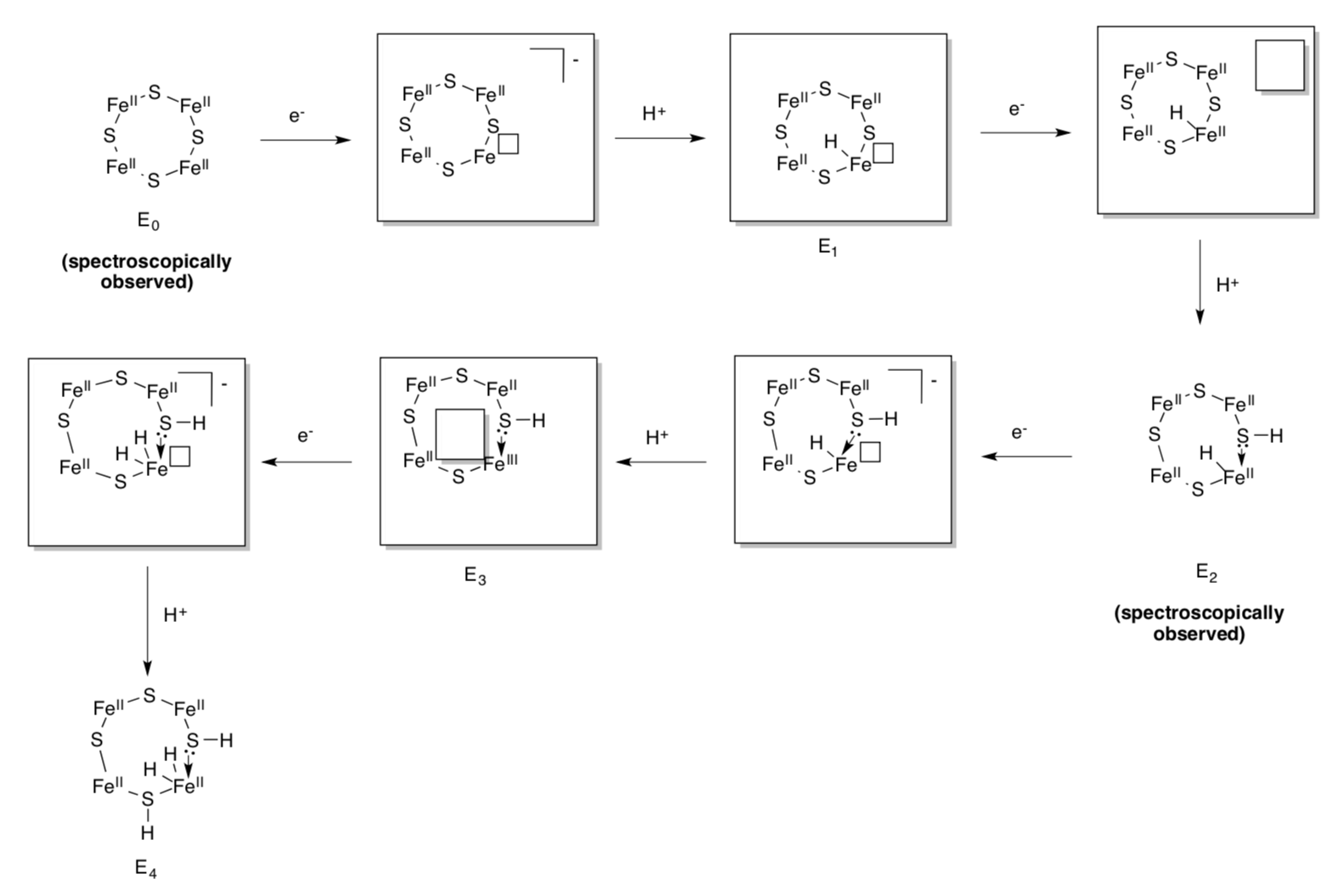
The state, E4, actually appears to have “bridging” hydrides. The iron-hydride bonds sigma donate into another iron. This feature may stabilize the hydrides in a resting state.
- Show what normally happens to hydrides in a protic environment (think of LiAlH4).
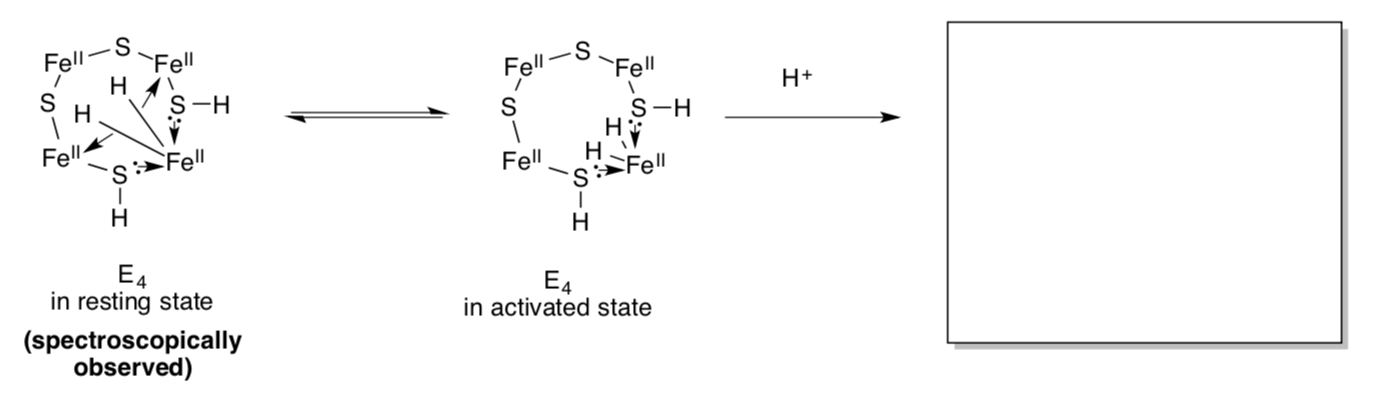
- Why might bridging make the hydrides less likely to get protonated?
Although state E4 appears to have a pair of hydrides, it does not undergo reductive elimination. However, H2 is produced in the presence of nitrogen.
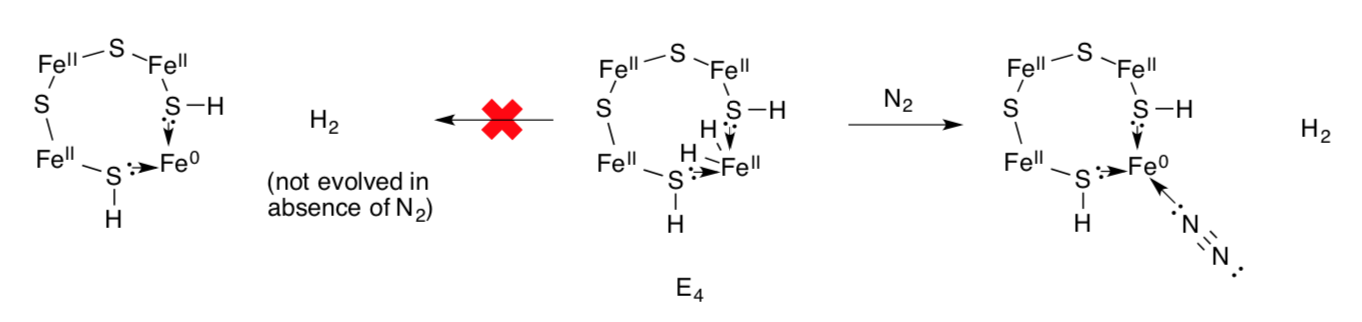
- Nitrogen is a σ-donor / π-donor / π-acceptor
- Why does reductive elimination make nitrogen binding easier?
- From an efficiency perspective, why might reductive elimination be prevented if there is no nitrogen present?
E4 is a crucial state because it binds nitrogen and initiates reduction.
- Fill in the missing items to complete the scheme.
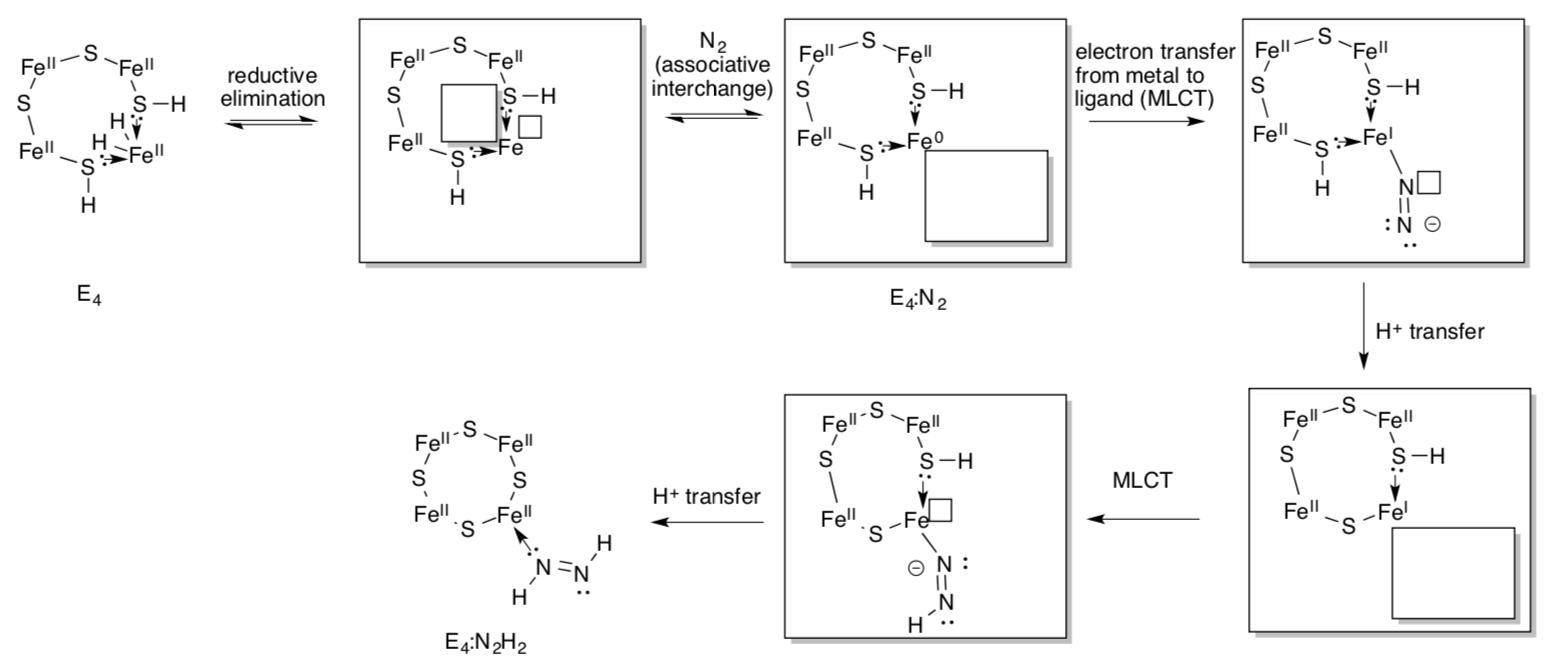
Note that the nitrogenous ligands are shown attached to just one metal. They may really bridge between two; researchers could not find conclusive evidence.
E4 is also capable of hydrogenating alkynes.
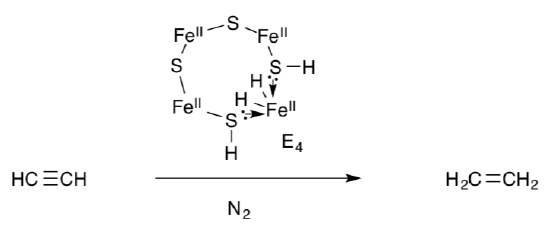
- Show the standard, three-step mechanism for alkyne hydrogenation using E4.
This mechanism inspired the following proposal for the remainder of the nitrogen fixation mechanism.
- Fill in the missing items.
Application Problem:
- In addition to performing the reaction N≡N → 2 NH3, nitrogenase is also capable of catalyzing the following reactions.
- Propose a catalytic cycle/mechanism (starting from intermediate E4 in the previous pages) for the reaction assigned by your instructor.
- HC≡CH → H2C=CH2
- N-=N+=O → N2 + H2O
- N=N=N- → N2 + NH3
- C≡N− → CH4, NH3, H3C–CH3, H2C=CH2 (CH3NH2)
- N≡C–R → RCH3 + NH3
- C≡N–R → CH4, H3C–CH3, H2C=CH2, C3H8, C3H6, RNH2
- O=C=S → CO+H2S
- O=C=O → CO+H2O
- S=C=N- → H2S + HCN
- O=C=N- → H2O+HCN,CO+NH3
- Propose a catalytic cycle/mechanism (starting from intermediate E4 in the previous pages) for the reaction assigned by your instructor.
- Nitrogenase contains two different classes of metal cluster: P-cluster ([Fe8S7]) and FeMoco ([MoFe7S9C·homocitrate]). The P-cluster is believed to mediate the electron transfer between the Fe protein and the MoFe protein via interconversions between its various oxidation states.
- Would this electron transfer be Inner Sphere Electron Transfer or Outer Sphere Electron Transfer?
- Explain your reasoning.
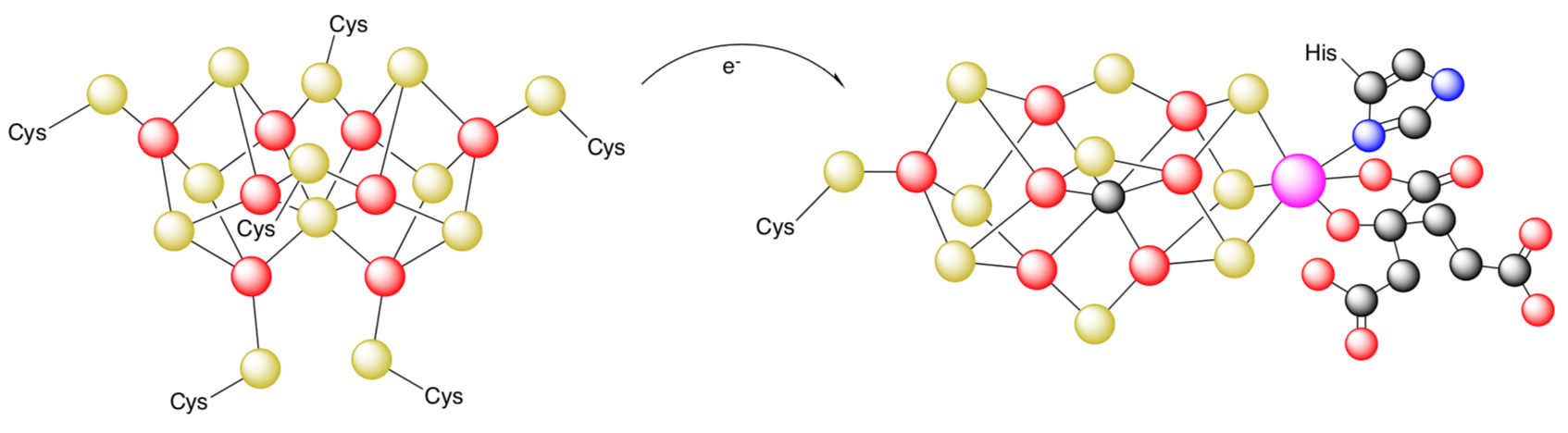
from Kresimir Rupnik, Yilin Hu, Chi Chung Lee, Jared A. Wiig, Markus W. Ribbe, and Brian J. Hales, J. Am. Chem. Soc., 2012 134 (33), 13749-13754.
- Vernon Gibson (Imperial) reported the following synthesis of an alkene polymerization catalyst (J. Am. Chem. Soc. 1999, 121, 8728). Show the product.
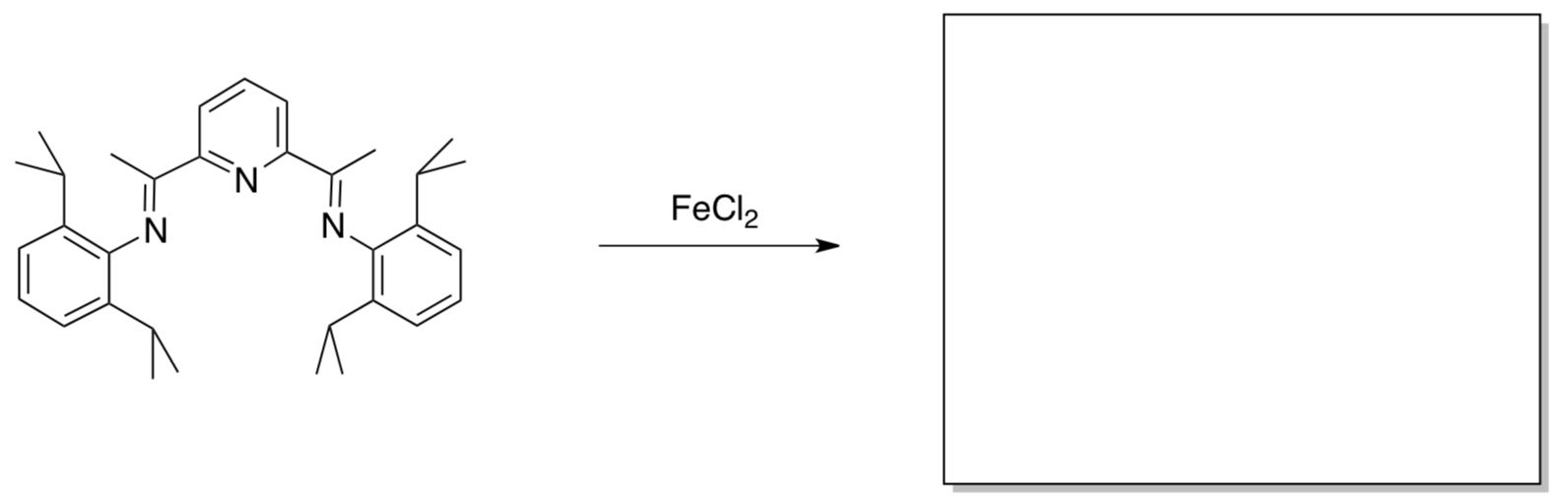
Paul Chirik (Princeton) thought this would be a good model for N2 activation. He treated the complex with sodium to obtain a bis(dinitrogen) complex (J. Am. Chem. Soc. 2004,126, 13794). Sandro Gambarotta (Ottawa) obtained the same product via addition of sodium hydride (Inorg. Chem. 2008, 47, 896).
Show the product of this reaction. Provide a mechanism (either route).
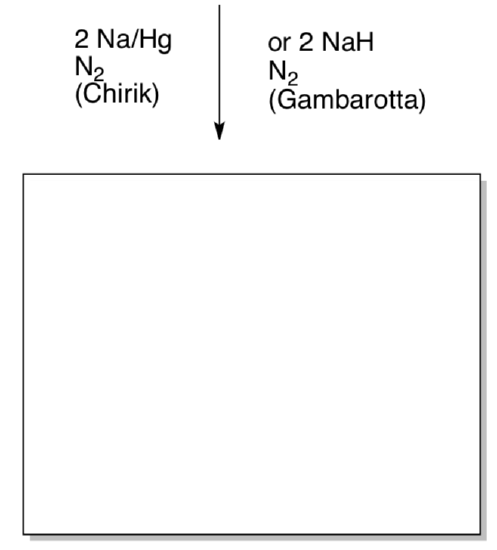
If the isopropyl groups are replaced with ethyl or methyl, a dimeric species with two terminal N2 and one bridging N2 is obtained instead. Show this complex.
The N-N distance in the bridging ligand is 1.123 A. The N-N distance in the terminal ligand is about 1.106 A. Explain the difference in bond lengths.
Why doesn’t a dimer result when isopropyl groups are used?

- Model Studies for Understanding Haber-Bosch
Evidence from simpler model compounds is often used both to completely elucidate the mechanism and to develop new catalysts. For example, about two dozen iron-dinitrogen complexes have been reported. Almost all have phosphine ligands. (Tyler, Coord. Chem. Rev. 2012, 1883-1894)
- Compared to other common donor atoms, such as oxygen or nitrogen, predict whether phosphine is a (circle one): strong sigma donor / weak sigma donor
- As a ligand, nitrogen is a (circle all that apply):
sigma donor | sigma acceptor | pi donor | pi acceptor
- Draw two possible structures of a nitrogen ligand attached to iron:
- bound by sigma donation (end-bound)
- bound by donation of pi bond (side bound).
- In general, we will always think of the nitrogen as end-bound. Add an additional resonance structure that shows stronger attachment to the iron.
- Draw two MO cartoon of a dinitrogen ligand attached to iron, showing both sigma and pi effects.
- Draw a d orbital splitting diagram of an Fe(II) in an octahedral environment and show how it changes when an N2 ligand binds.
- Why do phosphine donors stabilize iron dinitrogen complexes so well?
One example of a dinitrogen complex comes from Jonas Peters’ lab at CalTech,prepared via the following synthesis (Angew. Chem. Int. Ed. 2007, 5768):
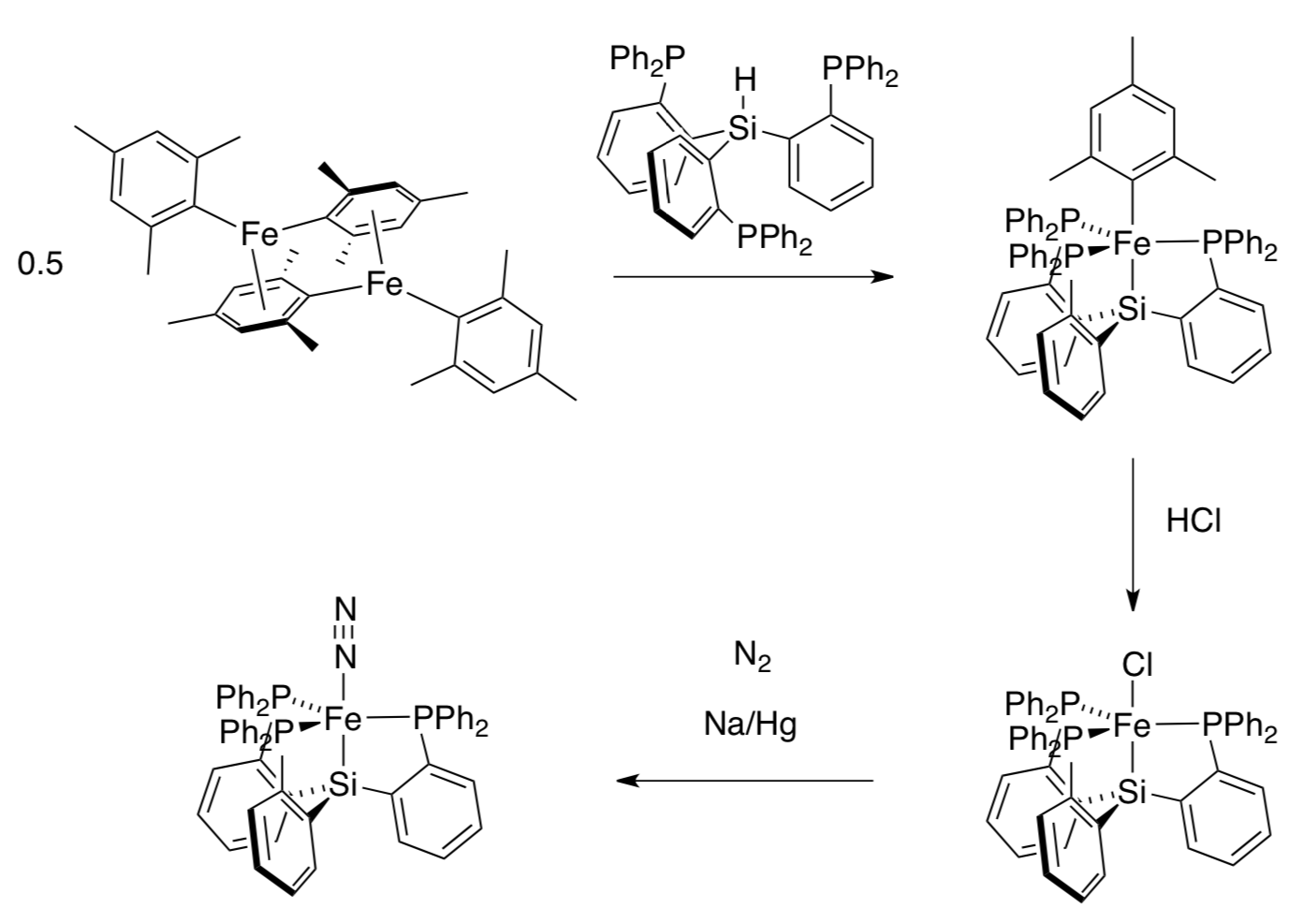
Reduction by sodium is a common method of obtaining reduced metal ions.
- Provide mechanistic details for the first two steps in the synthesis.
- Indicate oxidation states on the iron atoms.
- What is the side product of the third reaction?
- What did sodium give to iron in this third reaction?
Raman spectroscopy can be used to assess bond order: the weaker the bond, the lower the frequency of the corresponding Raman peak. The dinitrogen complex above has a N-N peak at 2008 cm-1. Addition of a second equivalent of Na/Hg reduces the frequency to 1967 cm-1.
- What is the role of sodium in this reaction?
- Why does the N-N frequency change when more sodium is added?
Addition of the strong acid HBF4 to the complex results in the production of hydrazine, H2N-NH2. (Peters, Inorg. Chem. 2009, 2507):
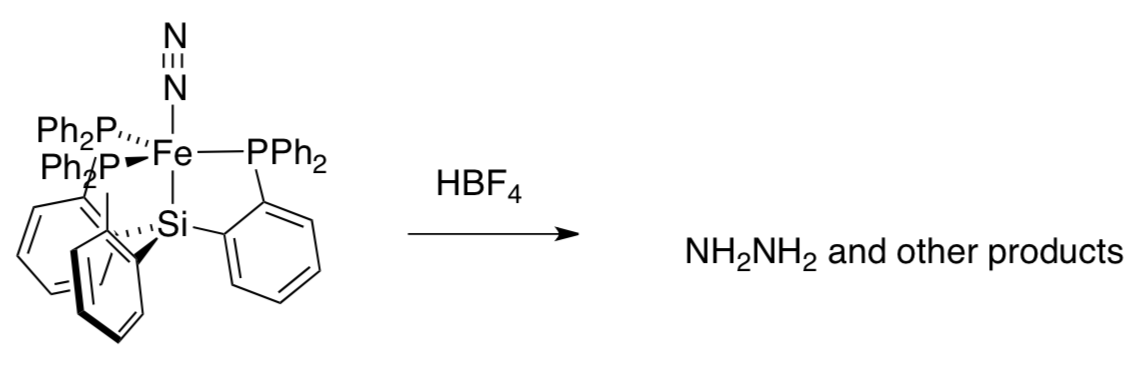
- State the significance of hydrazine production from this dinitrogen complex, in terms of modeling studies.
Sometimes, valuable insights can be gained by studying metals other than the one of direct interest in the real application.
A team led by Jean-Marie Basset, an organometallic chemist at Universite de Lyon, and Odile Eisenstein, a theoretical chemist at Universite de Montpelier, described the use of tantalum compounds on a silica surface to model the Haber-Bosch process (Science,2007, 317, 1056-1060).
Tantalum is partly useful in these studies because it forms much stronger bonds with nitrogen and oxygen than do most other transition metals.
- Provide one reason that Ta may form stronger bonds with nitrogen than Fe.
- Explain why strong bonds may be useful in modeling studies.
The researchers treated the silica surface with the Schrock alkylidene,tBuCHTa(CH2tBu)3, to obtain silica-bonded tantalum.
- Provide a possible product of the reaction.
- What is the side product?
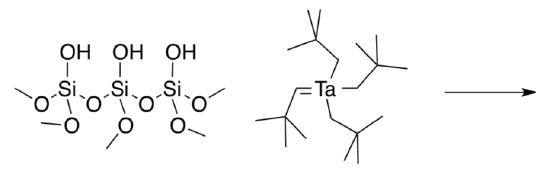
The remaining alkyl groups were removed via hydrogenolysis (treatment with H2), leaving tantalum hydrides bonded to the surface of the silica. Although reductive elimination in tantalum is rare, Eisenstein suggests that some Ta(III) may be in equilibrium with Ta(V) under these conditions.
- Draw the silica-supported tantalum hydrides that result after this step.
Nitrogen gas was introduced.
- Draw the surface N2 complexes that result after this step.
The researchers obtained IR and solid state 15N NMR spectra. Identical spectra were obtained by exposing the surface tantalum species with NH3 rather than N2.
- Use the table on the right to label peaks on the following (simulated) 15N NMR spectrum.
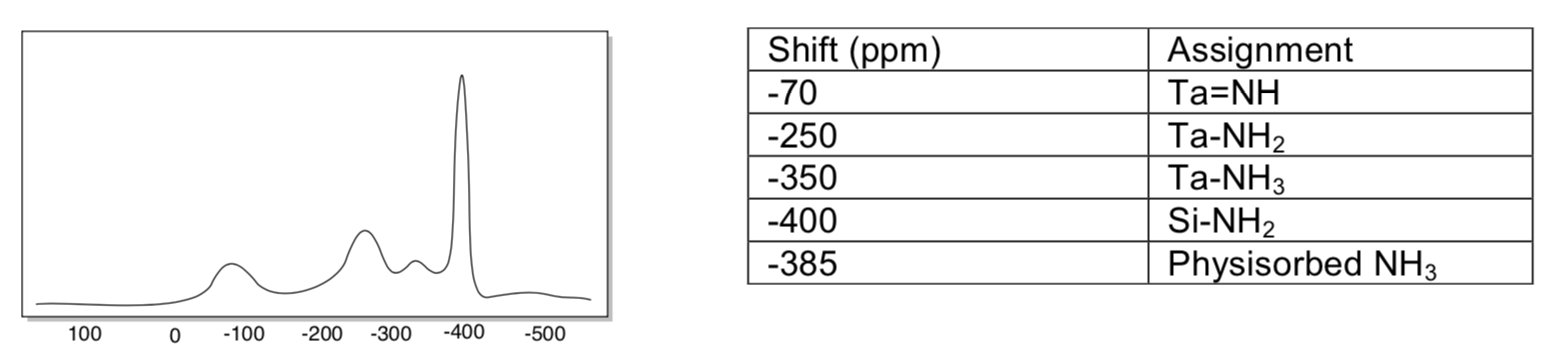
- A number of IR peaks were observed in the 3300-3500 cm-1 range. Interpret this data.
- Draw the species that formed on the surface, as indicated by the data.
- Why is it significant that similar spectra were obtained by using NH3 and N2?
- Model Studies for Understanding Biological N2 Fixation
Transition metal complexes are also used to model nitrogenase, the enzyme that converts nitrogen to ammonia in nature. Nitrogenase contains iron, bound by histidine, cysteine and sulfide donors. It reduces N2 via addition of protons and electrons.
- Compare and contrast the donating ability of phosphine donors used in iron / nitrogen complexes with the sulfur donors found in nitrogenase.
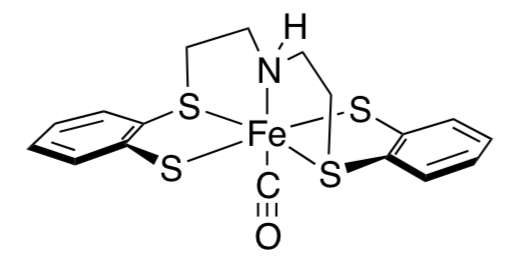
Dieter Sellmann at Friedrich-Alexander-Universitat Erlangen-Nurnburg has developed a number of models with sulfur-containing ligands, below. (Coord. Chem. Rev. 1999, 607- 627)
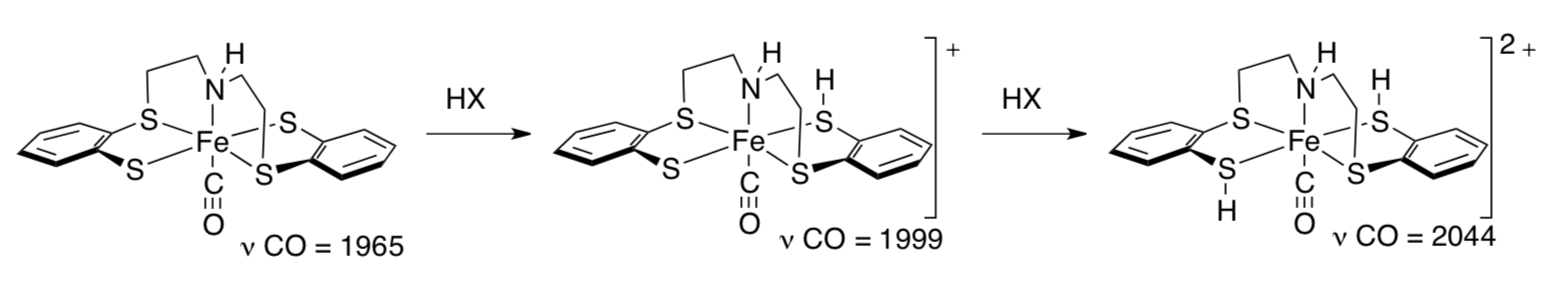
- Show the oxidation states on the iron atoms.
CO ligands are often used as probes of electron density at the metal. The more electron density, the weaker the CO bond in the ligand and the lower its frequency in the IR spectrum.
- Show why the CO bond strength depends on electron density at the metal.
- Why does protonation of the ligand affect the CO frequency in the IR spectrum?
- What happens to the Fe-C bond strength as the ligand gets protonated? Why?
- Sellmann was not able to make a dinitrogen complex. However, if he had, what would you expect to happen to the N-N stretching frequency as the ligands became protonated?
Model Studies for Understanding Biological N2 Fixation (cont.)
Electrochemical studies showed that the beginning compound above could not be reduced. However, the protonated compound could be reduced (-1340 mV) and the doubly protonated complex could be reduced twice (-540 mV and -1440 mV).
- Show the oxidation states on the iron atoms after each reduction.
- Why does protonation change how easily the complex is reduced?
Sellmann was unable to isolate a dinitrogen complex, but a hydrazine complex and a diazene complex were prepared as follows.
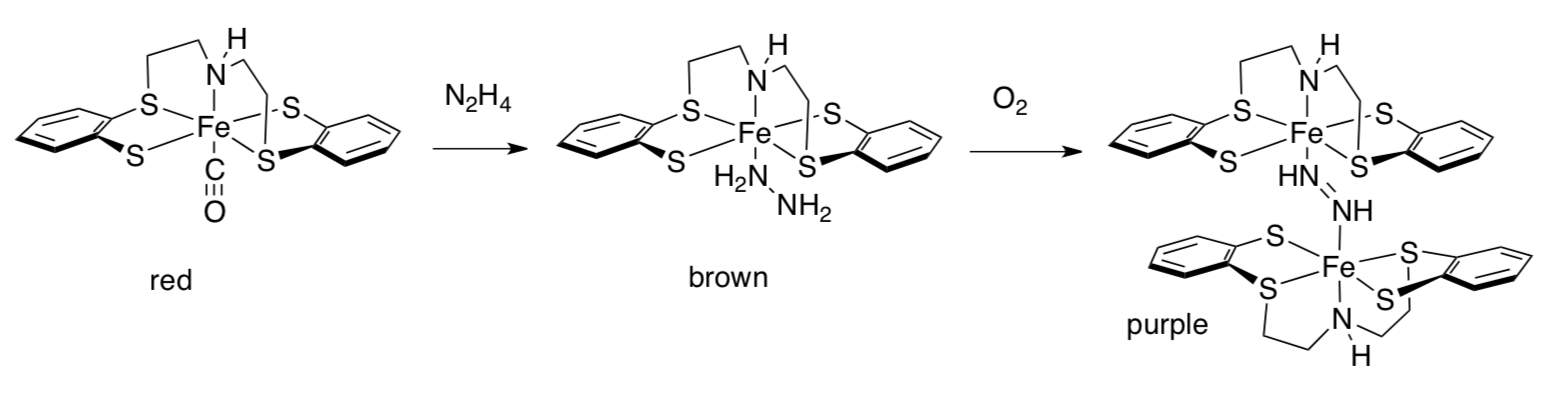
- Formation of diazene from hydrazine is an example of oxidation / reduction (circle one).
- Why does the diazene complex form a dimer? Draw an MO cartoon that shows how this structure is stabilized by conjugation.
Based on the crystal structure, Sellmann also believes there is a weak, stabilizing interaction between the diazene and the supporting thiolate ligands.
- Draw this interaction.
- Why isn’t it a stronger example of this type of interaction?
The diazene complex can be reversibly oxidized (+100 mV and +500 mV).
- Show what you think is happening during the electrochemical experiment.
- What would happen if the resulting complex were deprotonated?
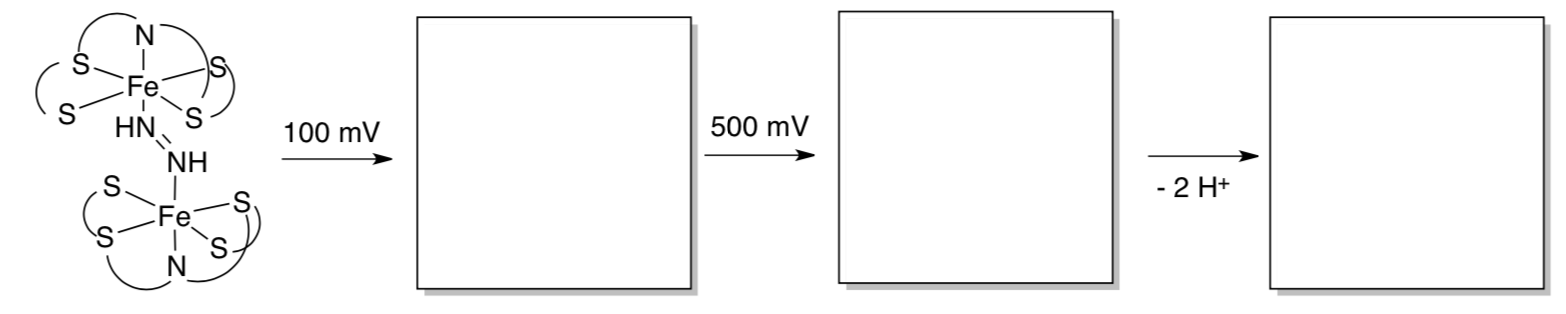
- Why is the reversibility of these steps significant in terms of modeling studies?


