25: Dynamic NMR
- Page ID
- 332830
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Dynamic NMR*
Reversible molecular processes which result in changes in chemical shift or coupling constant can be observed using dynamic NMR techniques. Typical systems that are exhibit changes include molecules undergoing conformational changes, proton exchanges, and ligand exchanges.
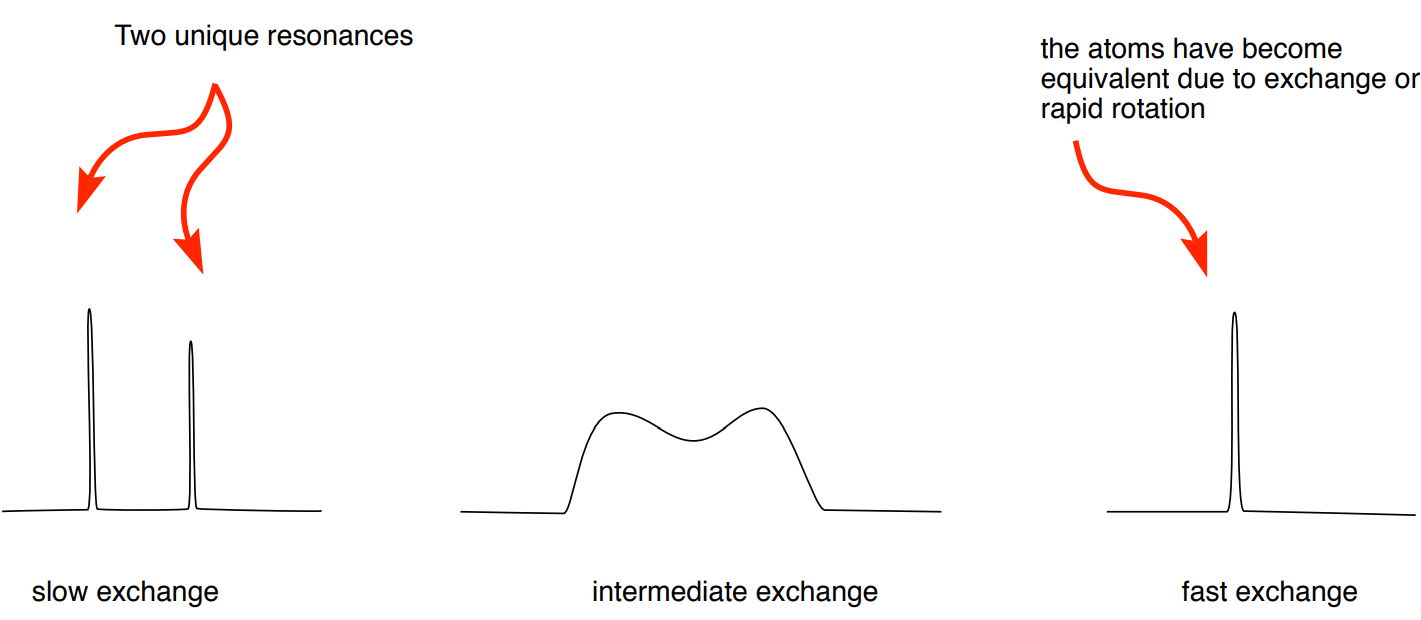
For two sets of signals to be recorded in an NMR experiment, exchange rate needs to be approximately 10-10,000 sec-1.
* All spectra are simulated.
Proton exchange on alcohols
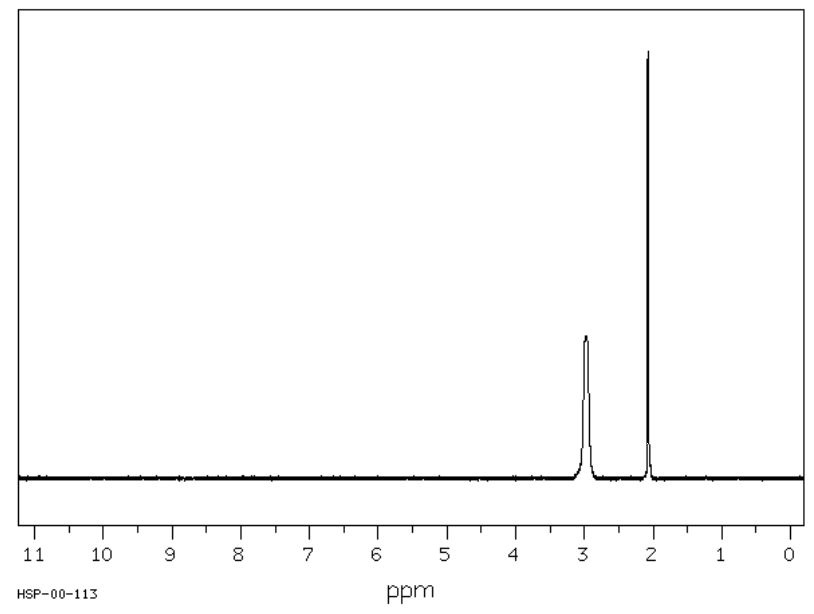
You have noticed this effect on alcohols and amines. Typically, in the 1H NMR, the peak corresponding to a hydrogen bonded to an O or N appears as a broad singlet. For example, the expected spectrum of \(\ce{CH3OH}\) is shown here.
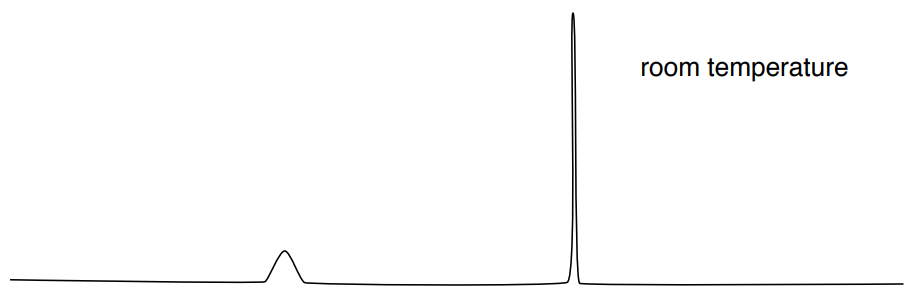
At room temperature, proton exchange on OH is fast (10-5 sec).

The rate of exchange can be changed by varying sample temperature; cooling slows exchange rate while warming increases exchange rates.
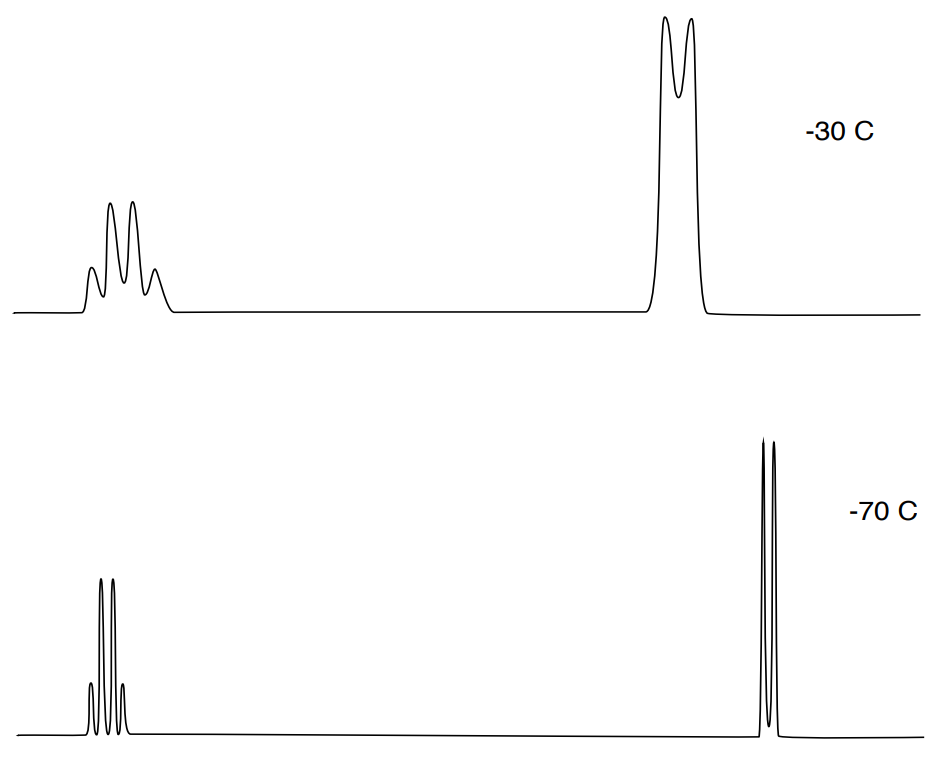
- Cooling __________ exchange rate while warming __________ rates.
- Comment on the change in coupling and chemical shift.
- Explain (in your own words) why the NMR coupling pattern is more distinct at cooler temperatures.
Hydrogen/Deuterium Exchange NMR
Protons that are slightly acidic will slowly exchange. If the sample is placed in D2O, the hydrogen atoms will be replaced with deuterium atoms.
- Draw a mechanism for the reaction of 1-propanol with D2O solvent molecules.
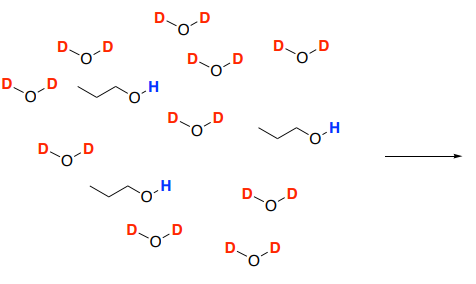
- Deuterium is not visible in the 1H spectrum so the OH peak will _________________ as the solution comes to equilibrium. Remember, in an NMR sample, the solvent greatly outnumbers the sample.
Deuterium exchange is a common technique used to examine protons are the amides in the backbone of a protein. The method gives information about the accessibility of various parts of the molecule, and thus the structure of the protein.
- If an amide is involved in an intramolecular hydrogen bond, it will be _________ [ more or less ] likely to exchange.
- If the amide is on the surface of a protein, it will be _________ [ more or less] likely to exchange.
- If an amide is in a site involved with protein-protein interactions, there would be it will be _________ [ more or less ] exchange processes in the complex vs the isolated protein.
- If conformation is altered as result of post-translational modification, enzyme activation, drug binding or other functional events, there will likely be _________ [a measurable / no measurable] change in the amount of exchange processes.
Restricted Rotation
Dynamic NMR has been used to study the restricted rotation around amide bonds, because of the importance of the phenomenon in peptide and protein structures and chemical behavior.
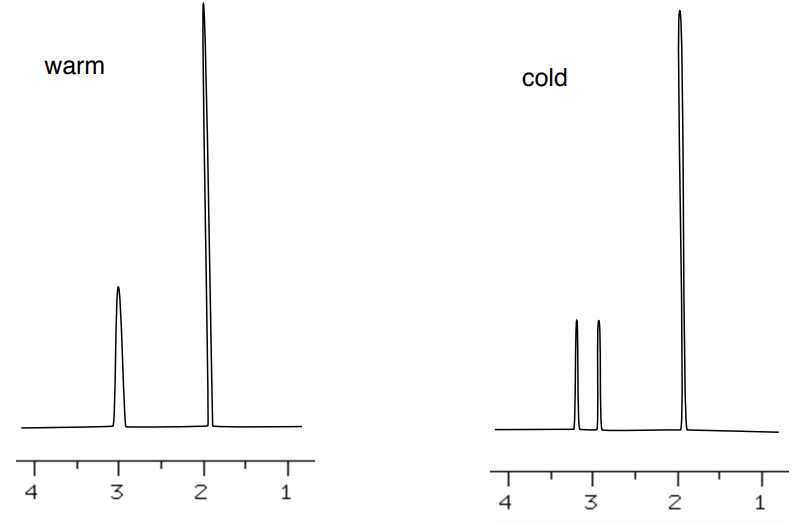
- Assign the peaks in 1H NMR of the N,N-dimethylacetamide.

There is restricted rotation around the CO-N bond.
- Draw a resonance structure to explain the restricted rotation.
Due to restricted rotation, spectrum changes at lower temperature.
- Explain why there are two peaks at lower temperature one broader peak at high temperature.
Conformational Changes
Cyclohexane chair conformers would be expected to would show the different sorts of protons (axial and equatorial) resonating at different frequencies, and so two signals would be seen, due to the protons being in slightly different chemical environments.
- Draw the two chair conformers of methyl cyclohexane.
However, only one signal can be seen as the two isomers are rapidly interconverting.
Similarly, 1,4-dioxane-d7 (shown below). Remember that d refers to deuterium (D) which is a heavier isotope of hydrogen.

- Draw the two chair conformers of 1,4-dioxane-d7.
- Draw the predicted 1H-NMR spectrum at room temperature.
- Explain why the 1H spectrum of 1,4-dioxane-d7 contains a peak at 3.7 ppm at room temperature but has two peaks at 3.6 and 3.8 ppm when the spectrum is obtained at 170K.
EXSY
The 2D EXchange SpectroscopY (EXSY) technique measures the exchange processes due to conformation, rotation or proton transfer in a 2D technique. EXSY is used to quantify dynamic processes in the 10–5000 ms time window. The method provides off-diagonal responses for spins that exchange slowly. Physical processes in this time window include slow conformational changes such as domain movement, ligand binding and release, interconversion of secondary structure and cis-trans isomerization.

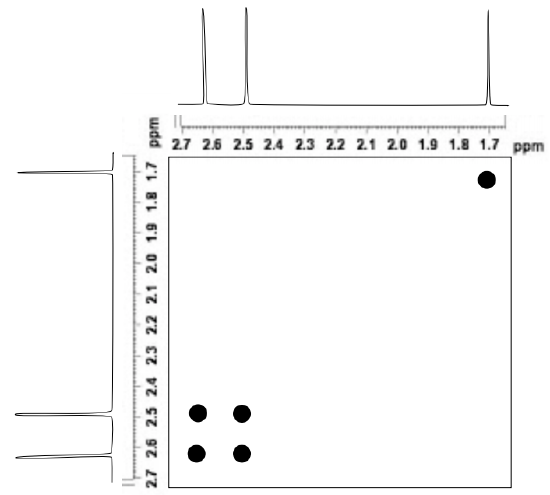
The figure below shows a simulated 2D 1H EXSY spectrum of N,N-dimethylacetamide at room temperature.
- Identify the cross peaks in this EXSY spectrum.
- Explain what these cross-peaks tell about this structure.
EXSY Application
The heptamethylbenzenonium ion undergoes an alkide shift that makes all seven methyl groups equivalent at sufficiently high temperature. Meier, B.H.; Ernst, R.R. J. Am. Chem. Soc., 1979 101, 6442-6443.
- Draw a few of these 1,2 alkide shifts.
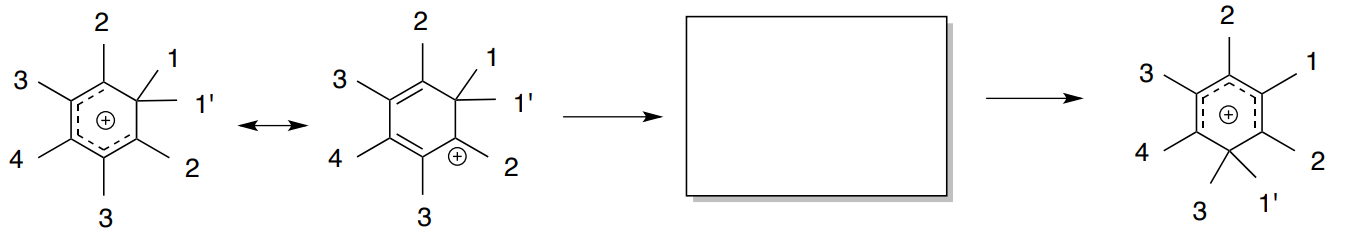
There has been some discussion whether this shift involves either a 1,2 methide shift or a random shift with jumps between all possible positions.
A simulated EXSY NMR spectrum of this equilibrium process.

- Does this spectrum indicate that the shift is occurring as 1,2 shifts or random jumps? Explain.
Saturation Transfer Difference NMR
Saturation Transfer Difference (STD NMR) detects transient binding of small molecule ligands to macromolecular receptors.
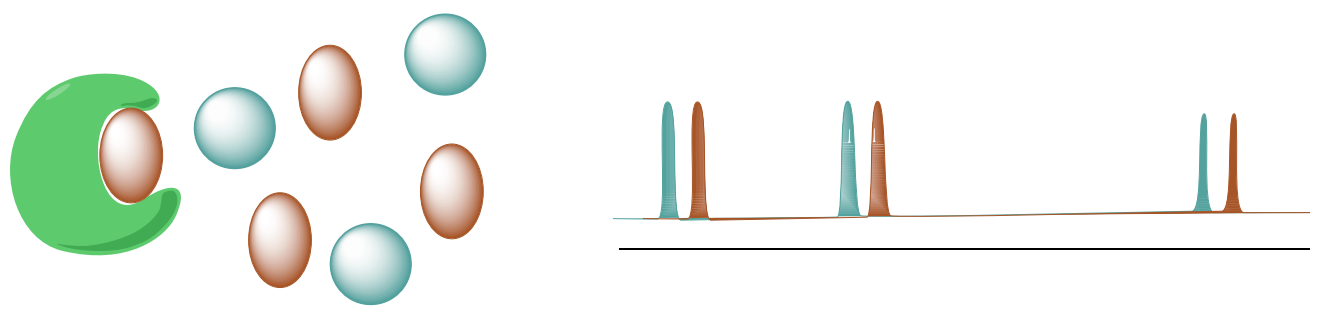
A. STD selectively saturates protons of the macromolecular receptor by irradiating the spectral region containing resonances of the macromolecule but NOT any peaks from the possible ligands.
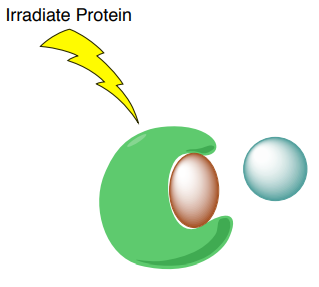
B. The saturation will transfer to the nuclei of the ligands that are bound the receptor. This saturation transfer increases the intensity of the NMR signals from the ligands that are binding to proteins.

Range of applicable dissociation constants is approximately 10-3 - 10-8 M.
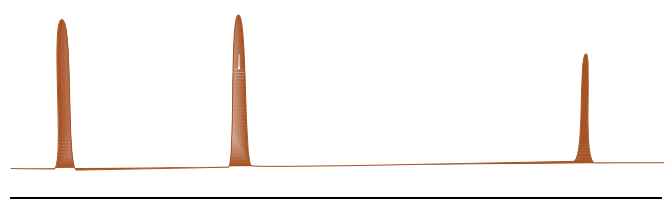
C. Subtraction of resulting spectrum from the reference spectrum without saturation shows only signals of the binding ligands.
STD Screening Tool
This experiment is often used as a screening tool to determine which compound binds to the protein in the context of drug discovery.
Cabrita, et. al., J. Chem. Educ. 2011, 88, 990–994.
Human serum albumin (HSA) is used as a receptor and 6-D,L -methyl-tryptophan (6-CH3 -Trp) and 7-D,L methyl-tryptophan (7-CH3 -Trp) as the ligands.
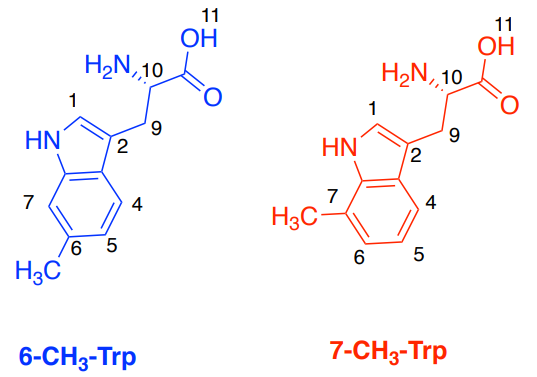
A. Simulated Spectra of the two ligands: 6-CH3 –Trp (blue) and 7-CH3 –Trp (red).
B. STD Spectra with HSA and two ligands
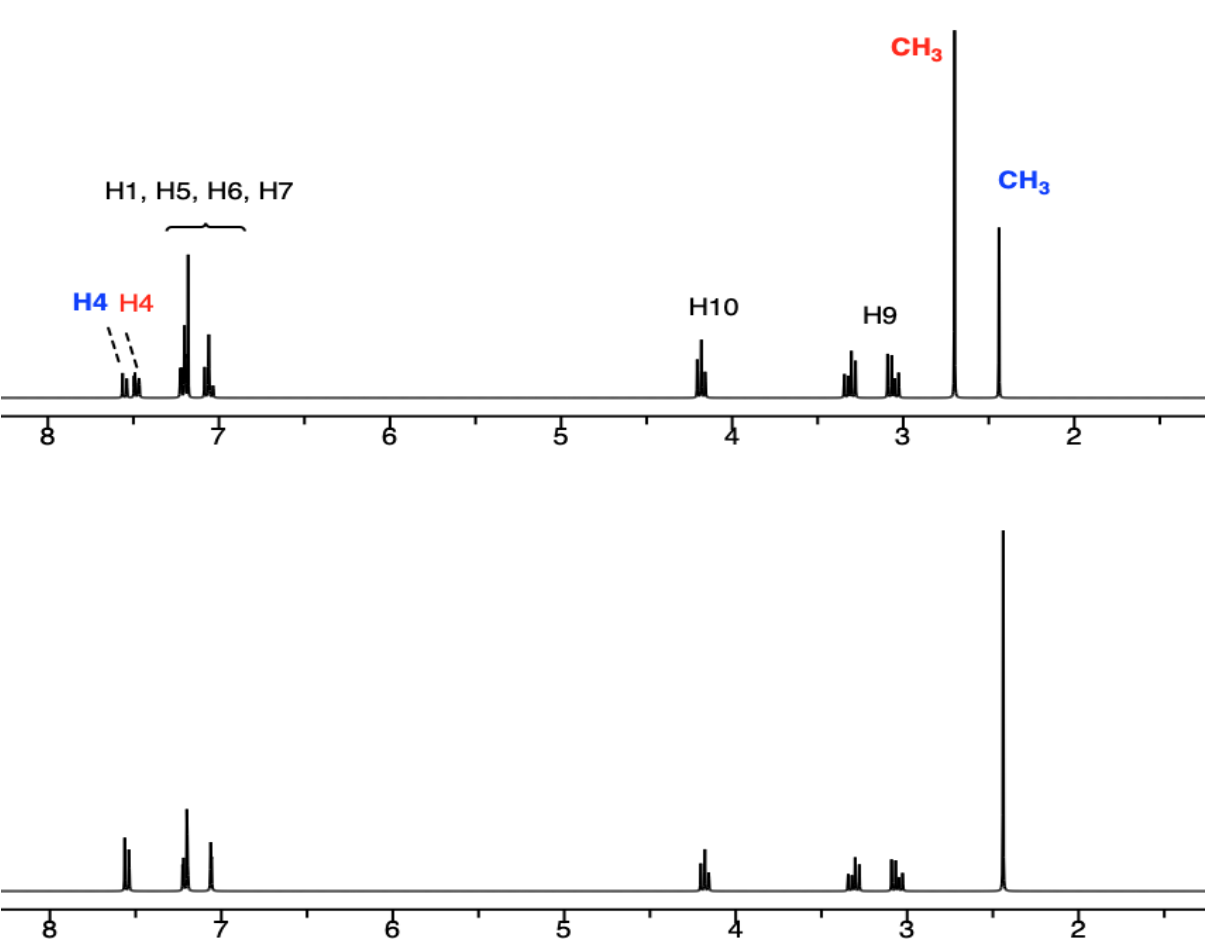
- Which ligand is binding to the active site of HSA?
6-CH3 –Trp OR 7-CH3 –Trp
- Which portion of the ligand appears to be binding?


