Enzymes and Catalysis
- Page ID
- 50922
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The most efficient and amazing catalysts of all are the biological molecules known as enzymes. One of these, nitrogenase, is found in algae, bacteria, and microorganisms associated with leguminous plants such as soybeans. Nitrogenase can fix nitrogen from the atmosphere at room temperature (instead of at 500°C as in the Haber process). It does so efficiently enough that only a few kilograms of enzyme are necessary to fix about 44 × 1012 g of nitrogen worldwide each year. Other enzymes carry out other reactions with comparable efficiencies.
All enzymes are large protein molecules whose molar masses exceed 20 000 g mol–1. They are condensation polymers of amino acids, and a typical section of a long protein chain may be represented as
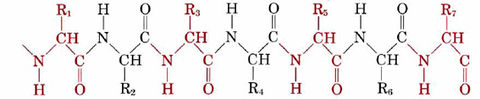
The alternate red and black segments each represent a different amino acid, and the symbols R1, R2, R3, etc., stand for different organic groups called side chains. In the amino acid glycine, for instance, the side chain is a hydrogen atom, while for glutamic acid it is the group —CH2—CH2—COOH. A typical enzyme molecule contains as many as 20 different kinds of amino acids, while the chain may be as long as 200 or 300 amino acid units. (More details on the amino acids, their side chains, and the primary, secondary, and higher order structures of proteins are given in their respective sections.)
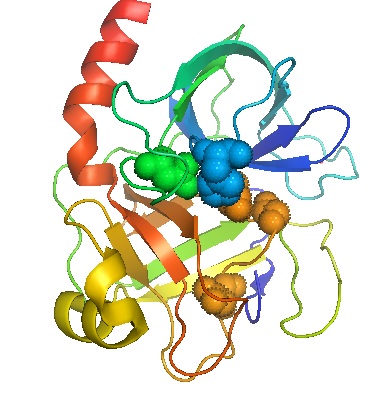
Figure \(\PageIndex{1}\) The complex folding of the chain of amino acids in the enzyme trypsin. Amino acid residues important for the catalytic activity of the molecule centers are shown as filled in spheres. Accordingly this region is called the active site.
The polymer chain of amino acid units in an enzyme is usually wrapped around itself in a complex fashion to form an overall shape which is approximately spherical. The conformation of the protein chain in the enzyme trypsin has been determined experimentally and is shown in Fig. \(\PageIndex{1}\). The chain appears quite random, almost as though it were a ball of tangled string or yarn, but there are certain molecular features in the region shown in color in the diagram which are essential to its operation as a catalyst. Accordingly, this region of the molecular surface is called the active site.
An important feature of the catalytic behavior of enzymes is their specificity. Most enzymes are able to catalyze only one particular chemical reaction or at most only a few closely related reactions. Some interesting examples of specificity involve the enzymes which, as part of the process of digestion, catalyze the breakdown of other proteins into their constituent amino acids in the stomach and intestines. These enzymes all catalyze hydrolysis of the C—N bond in the amino acid chain according to the following equation:
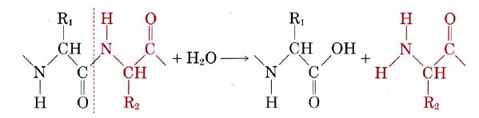
When all the C—N bonds in a protein are broken, it is transformed entirely into amino acids. Amino acids can penetrate the walls of the intestine and be utilized by the body to build up other protein chains.
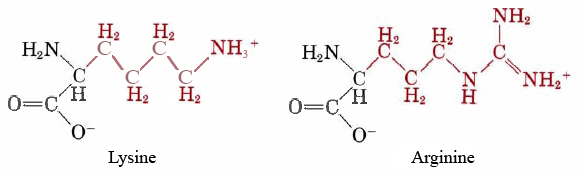
Hydrolysis of this type of C—N bond is also catalyzed by acids and bases. Thus, for instance, a standard laboratory method for breaking up a protein into its constituent amino acids is to heat it overnight at 100°C in 1 M HCl. Despite the higher temperature, this acid-catalyzed hydrolysis is much slower than one catalyzed by an enzyme, and all the C—N bonds are broken, no matter what amino acids are involved. When an enzyme is used, by contrast, only certain C—N bonds are hydrolyzed. The enzyme carboxy-peptidase will only catalyze the hydrolysis of a C—N bond at the end of an amino acid chain. On the other hand, trypsin will only operate if one of the amino acids involved in the bond is lysine or arginine.
Enzymes usually behave very simply from the point of view of chemical kinetics. The rate law is usually found to be first order with respect to the enzyme concentration and also first order with respect to the substrate (the substance on which the enzyme acts). In other words
\[\text{Rate}= k(c_{enzyme})(c_{substrate})\]
Thus the activated complex consists of a combination of one enzyme molecule and one substrate molecule. Since the enzyme molecule is usually much larger than the substrate molecule, we can think of the substrate molecule as being held down on the surface of the enzyme and enzyme catalysis as being related to heterogeneous catalysis.
Using multiple techniques, such as X-ray crystallography to determine three-dimensional structures of enzymes, as well as experimental kinetics, biochemists can understand the finer details of how an enzyme is able to operate so efficiently as a catalyst. Among the best understood is the enzyme trypsin, which, as we have just mentioned, catalyzes the hydrolysis of C—N bonds attached only to the amino acids lysine and arginine. A somewhat oversimplified and schematic account of the operation of this enzyme is given in Fig. \(\PageIndex{2}\).
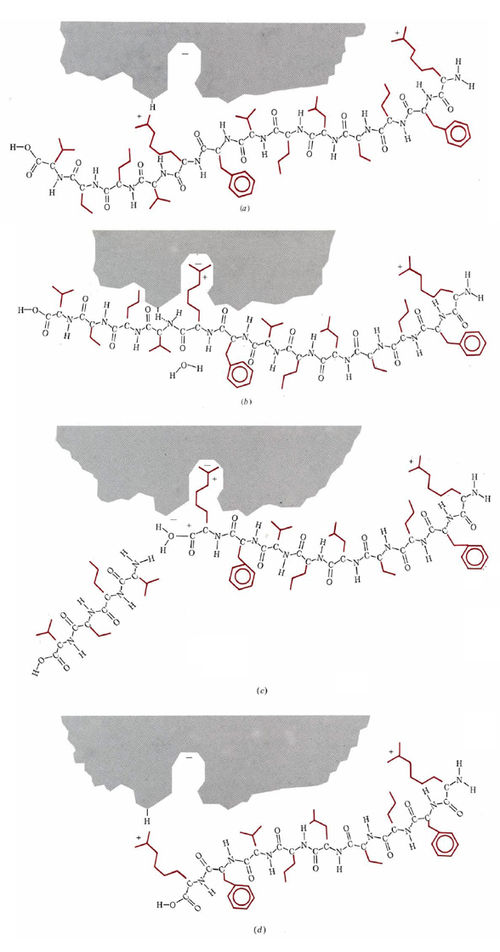
Figure \(\PageIndex{2}\) A schematic description of the operation of the enzyme trypsin in hydrolyzing a protein chain. (a) The protein chain approaches the active site, and an arginine side chain is attracted toward the pocket. (b) The arginine side chain lodges in the pocket, while a conveniently situated acidic proton bonds to the N atom of the C—N bond which is about to be hydrolyzed. (c) The proton catalyzes the breaking of the C—N bond, and one fragment of the C—N chain moves away. An H2O molecule can now enter and bond to the positively charged C atom. (d) The H2O molecule has added an OH group to the end of the chain and replaced the lost proton on the enzyme. The arginine side chain is released from the pocket, and the enzyme is ready for the next customer.
The active site on the surface of the enzyme, shown in gray, contains an opening or pocket which is just large enough to accommodate the long side chains of lysine or arginine, and which also contains a negative ionic charge deep inside it. Side chains belonging to other amino acids are the wrong size or shape or are not positively charged, and so they cannot be held in this pocket, but an arginine or lysine side chain can be. Once the side chain is held by the pocket, there is an acidic hydrogen atom on the enzyme in exactly the right position to protonate the nitrogen atom in a C—N bond in the protein chain. The enzyme thus functions as an acid catalyst for the hydrolysis of the C—N bond. It is, of course, much more efficient than a nonen-zymic catalyst such as HCI. Partly this is because the proton is in exactly the right place to donate once the side chain has entered the pocket. Partly, also, it is because there is considerable lowering in the activation energy due to the ionic attraction between the enzyme and substrate.
From ChemPRIME: 18.10:Catalysis
Contributors and Attributions
Ed Vitz (Kutztown University), John W. Moore (UW-Madison), Justin Shorb (Hope College), Xavier Prat-Resina (University of Minnesota Rochester), Tim Wendorff, and Adam Hahn.


