12.P: Problems for Chapter 12
- Page ID
- 975
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Link to Solution Manual
P12.1: A bacterial enzyme called aminoglycoside aminotransferase is thought to be responsible for resistance to a class of antibiotics that includes apramycin, ribostamycin and paromomycin. The enzyme catalyzes an acylation of the antibiotic compound, as shown below for apramycin.
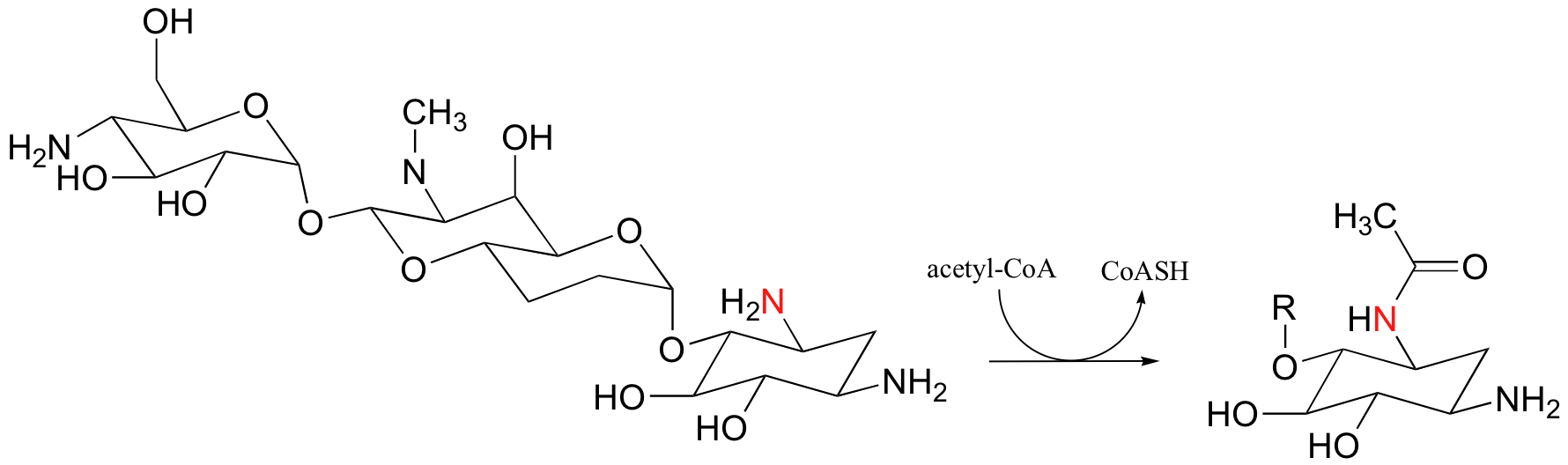
Show a mechanism for this reaction (use an 'R' group to abbreviate the antibiotic structure, as shown in the product drawing above).
P12.2: Propose a mechanism for the following reaction:
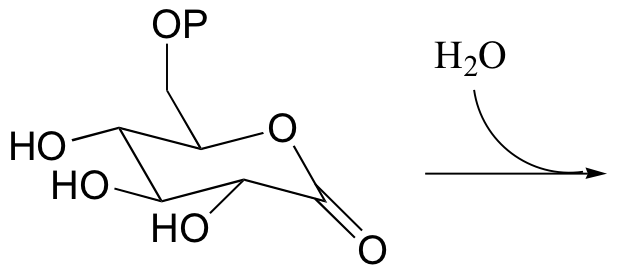
P12.3: Show the product of the hydrolysis of the lactone (cyclic ester) group in the molecule shown below, and provide a mechanism.
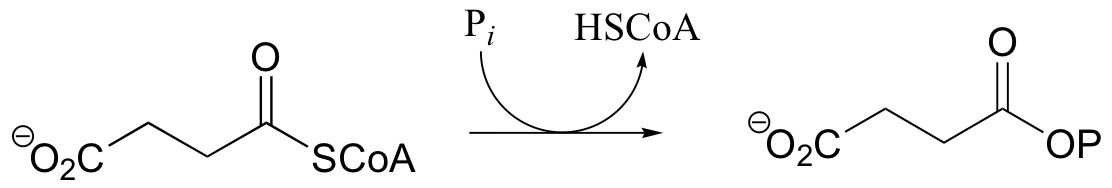
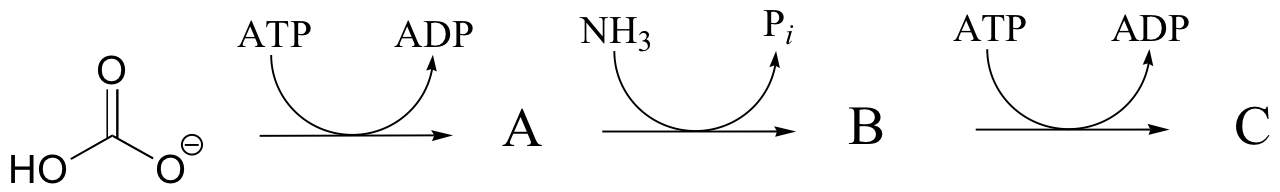
P12.4: Predict the structures of compounds A, B, and C in the reaction pathway below. Compound C contains an activated carboxylate functionality. Use abbreviations as appropriate.
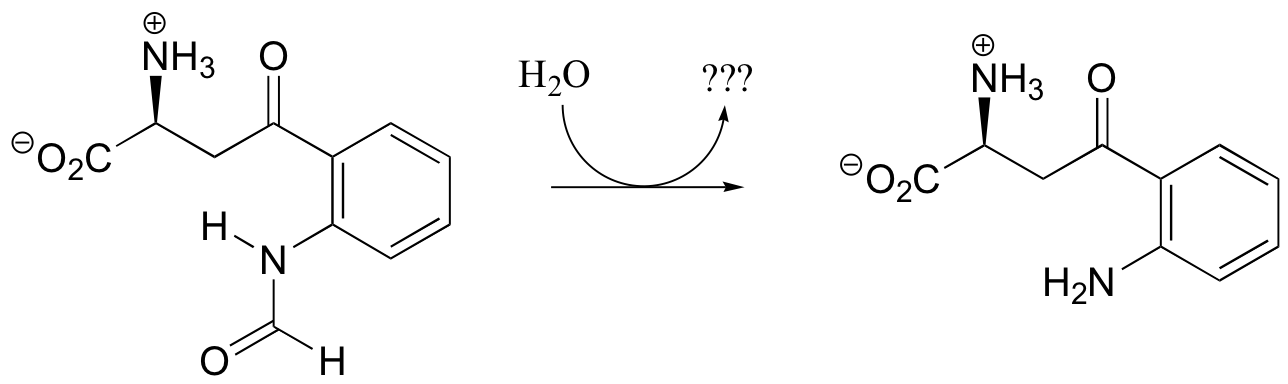
P12.5: Draw the structure of the missing product (designated below by question marks) in the reaction below. How would you describe this reaction inorganic chemistry terminology?
P12.6: Look back at the second (hydrolysis) step of the epoxide hydrolase reaction discussed in section 9.4A (the asterisk denotes an 18O isotopic label):
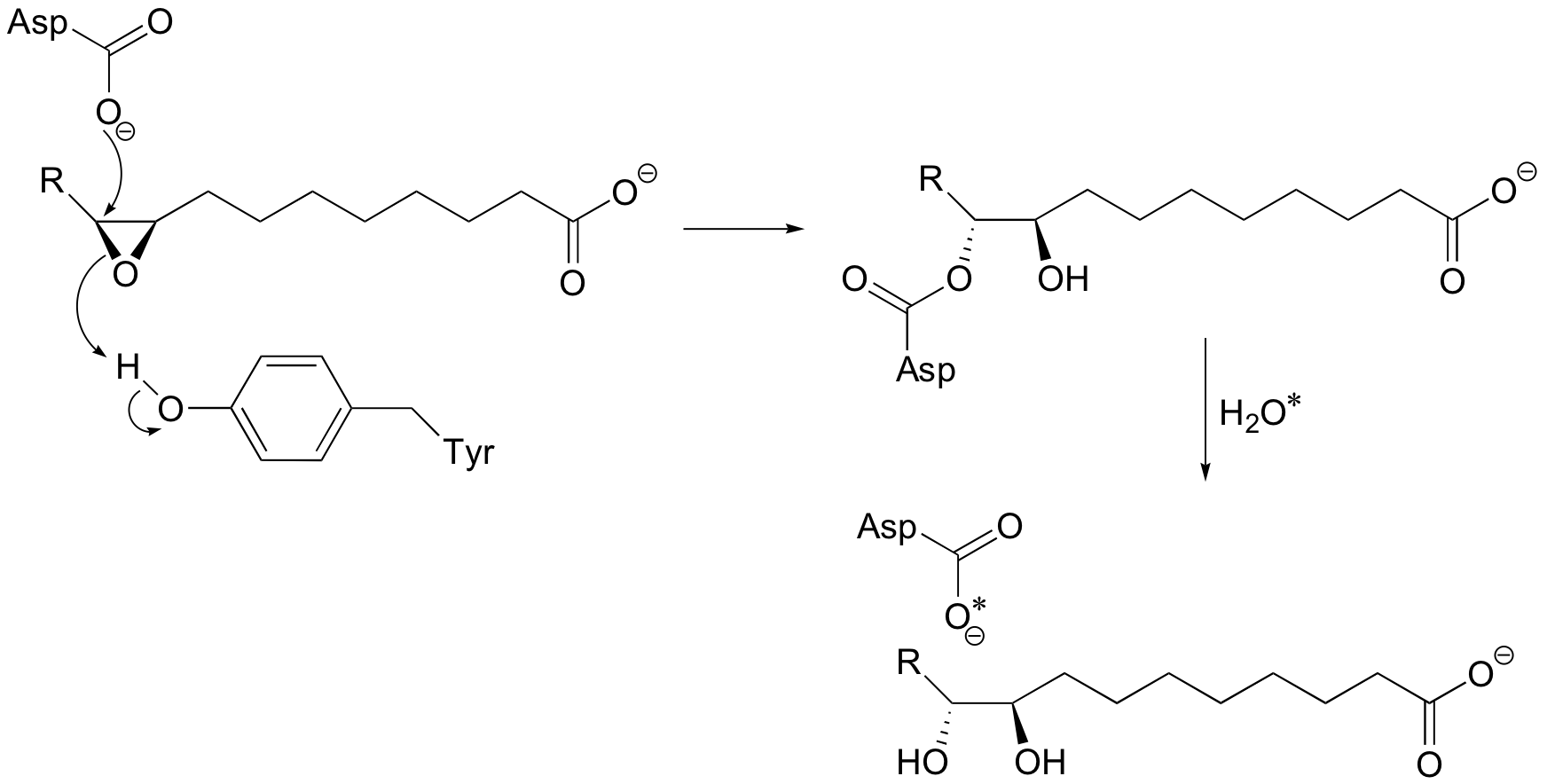
Write a mechanism for the second (hydrolysis) step. Can you see now why this step results in retention of configuration at the epoxide stereocenter?
P12.7: A different epoxide hydrolase (from the one mentioned in problem 12-6) catalyzes the reaction shown below. When the reaction was run with enzyme in which the active site aspartate was labeled with 18O, the label was incorporated into the product (see J. Am. Chem. Soc. 115, 10466).
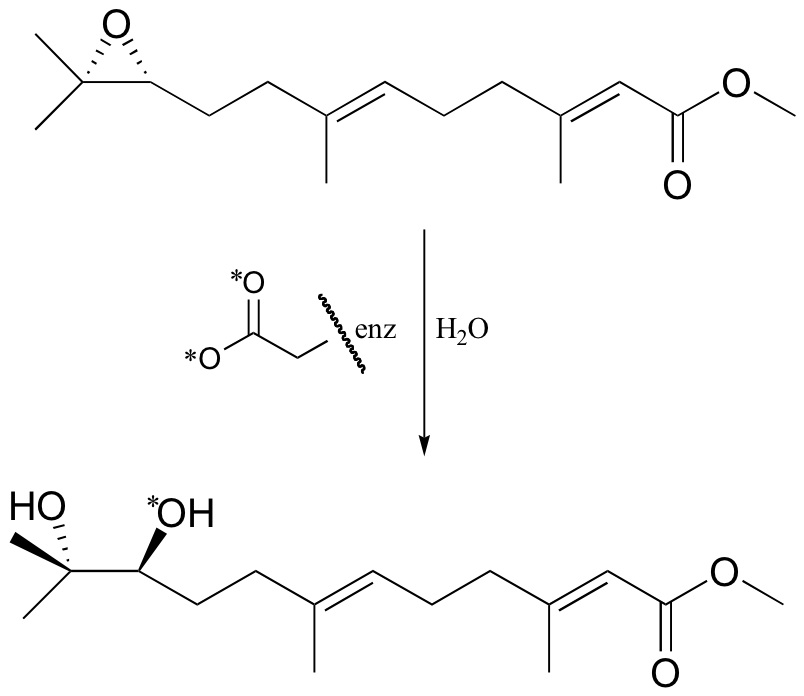
a) Does the evidence indicate that this reaction proceeds with a similar mechanism to the one above, in which an active site aspartate serves as the ring-opening nucleophile? Explain.
b) Provide an alternate mechanism which, if it were operating, would result in an unlabeled product with the opposite stereochemistry.
P12.8: Propose a mechanism for the following reaction:
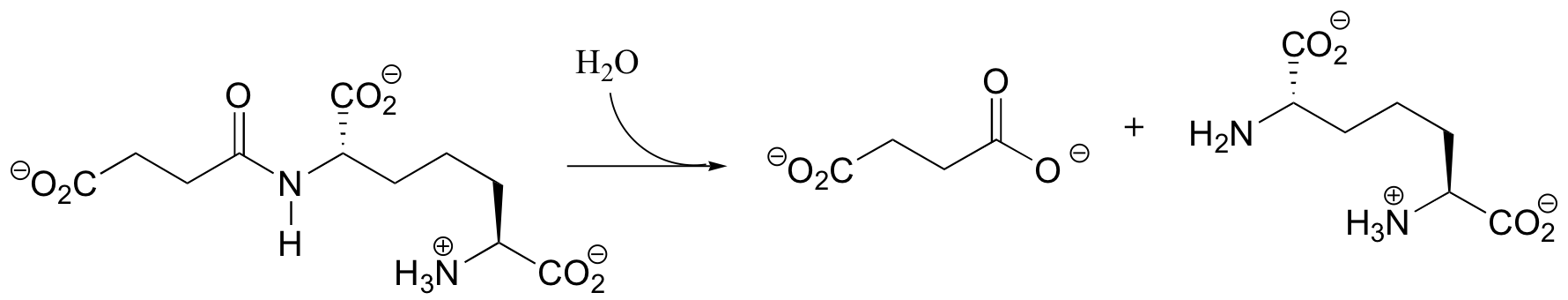
P12.9: Before fatty acids are transported across the inner mitochondrial membrane, they are temporarily attached to a transport molecule called carnitine.
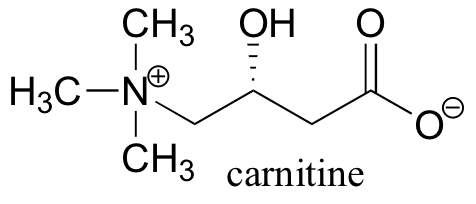
Fatty acids are linked to carnitine via an ester linkage. Draw the structure of fatty acyl carnitine (use R to denote the fatty acid hydrocarbon chain), and provide a complete mechanism for its formation (begin with fatty acyl-SCoA).
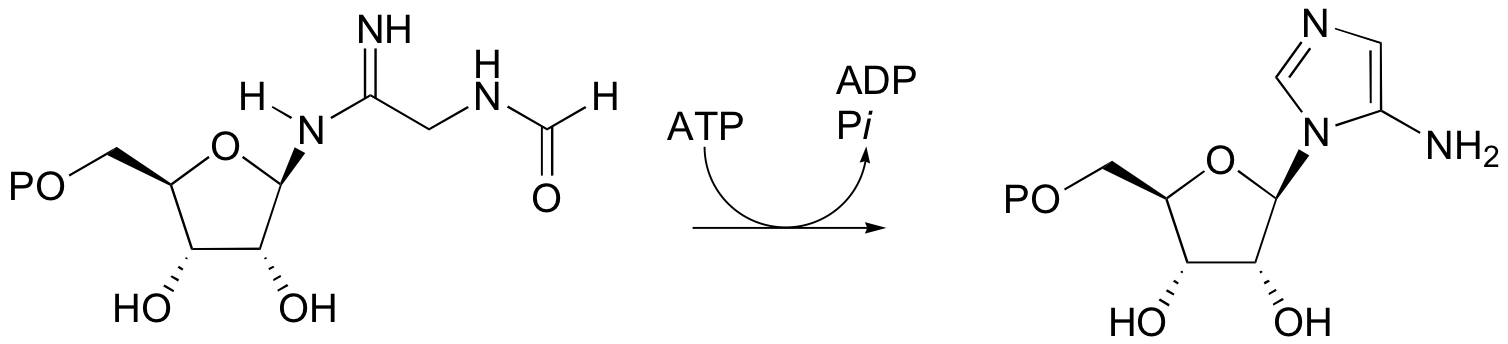
P12.10: The following reaction is a step in the biosynthesis of purine ribonucleotide monophosphates (AMP and GMP). The mechanism is similar to that of argininosucccinate synthase. Provide a complete mechanism.
P12.11:
a) Propose a mechanism for the following reaction.
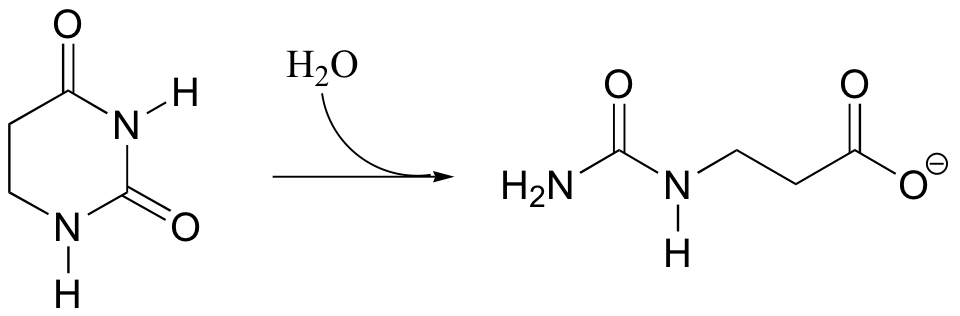
b) The substrate in this reaction could also undergo an alternate hydrolysis reaction, leading to a different product. Draw the structure of this alternate, hypothetical product.
c) Hypothesize as to how the enzyme, in catalyzing the reaction shown in part a of the problem, might direct the hydrolysis in the 'correct' regiochemical course.
P12.12: Propose a mechanism for the following reaction.
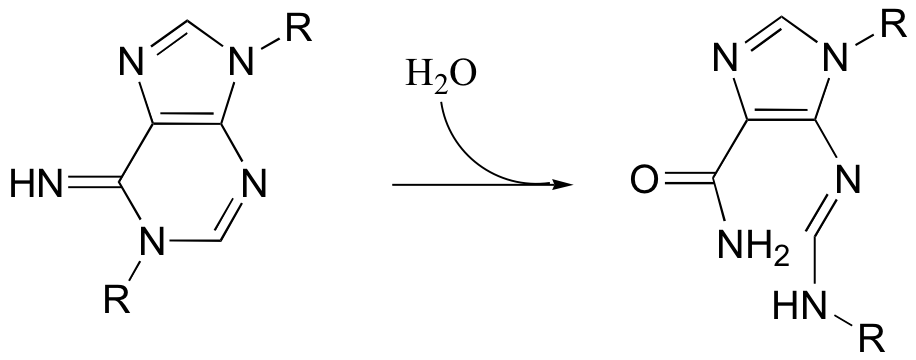
P12.13: Propose a mechanism for the following reaction.
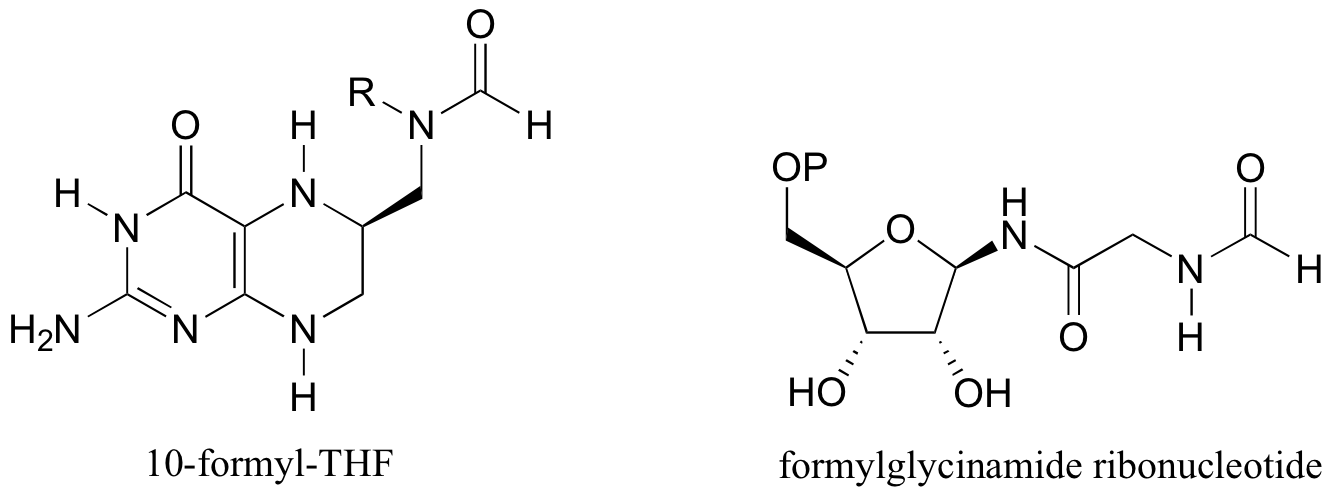
P12.14: The coenzyme tetrahydrofolate (THF) participates in single-carbon transfer reactions. One derivative, called 10-formyl-THF (abbreviated structure shown below), transfers a formyl group early in purine ribonucleotide biosynthesis to form formylglycinamide ribonucleotide. Draw the predicted structure of glycinamide ribonucleotide, the substrate for this reaction. Also, draw the structure of THF, the other product of this reaction.
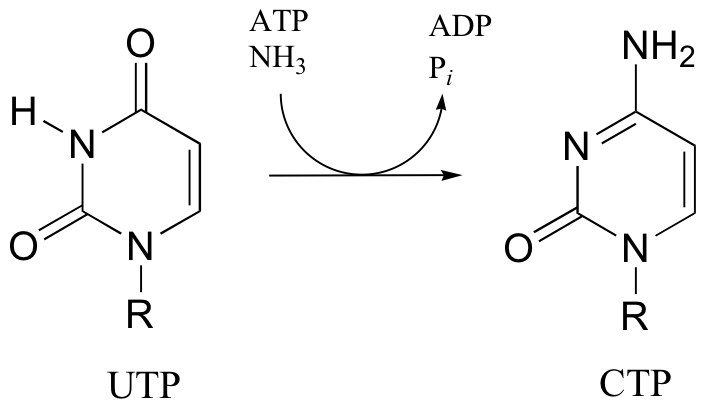
P12.15: One of the key steps in the biosynthesis of purine nucleotides (guanosine and adenosine) is shown below.
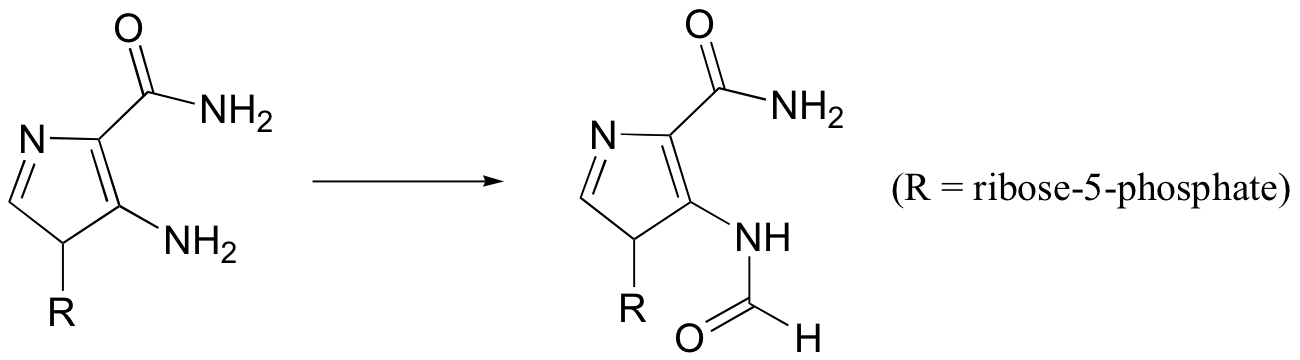
In eukaryotes and bacteria, this reaction requires the coenzyme 10-formyltetrahydrofolate (see structure in problem 12-14). In archaeal species, however, the same reaction requires only free formate and ATP. Suggest reasonable mechanism for the archaeal version of this biosynthetic transformation.
P12.16:
a) Propose a mechanism for the following reaction:
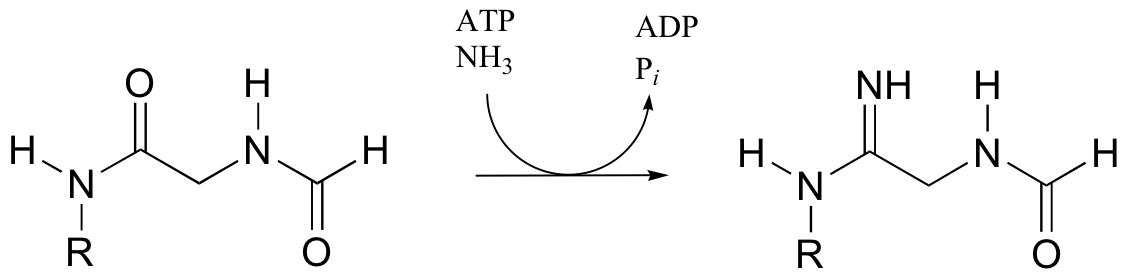
b) In the final step of the urea cycle (in amino acid degradation), arginine is hydrolyzed to urea and ornithine. Propose a mechanism.
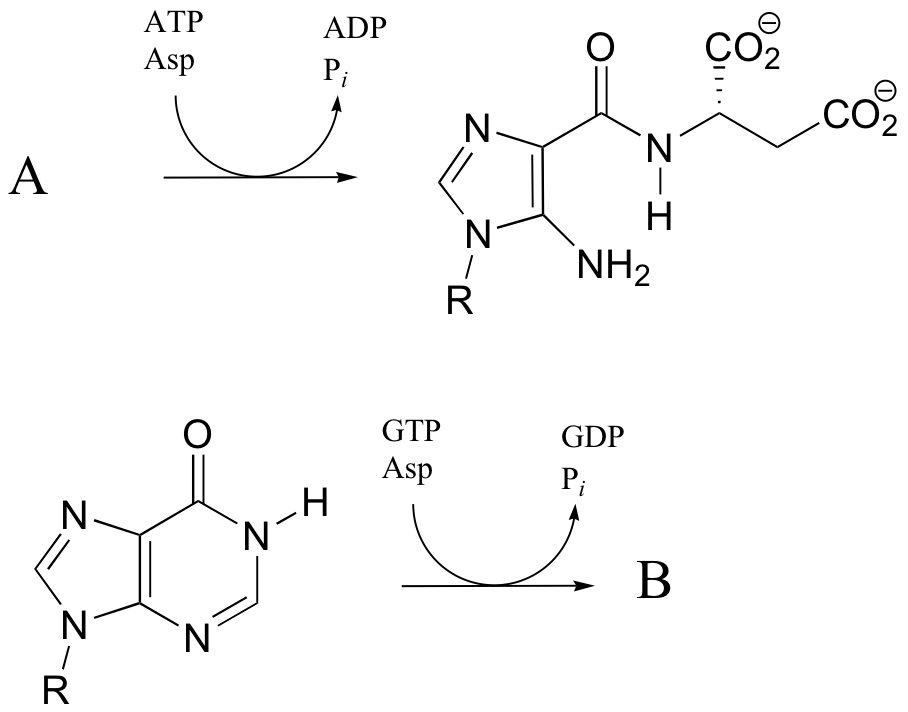
P12.17: The reactions below are both part of nucleotide biosynthesis, and both involve the condensation of aspartate with a second molecule. Predict the structures of compounds A and B. Compound A contains a carboxylate group. The reaction that leads to compound B is very similar to that catalyzed argininosuccinate synthetase (discussed in section 12.6).
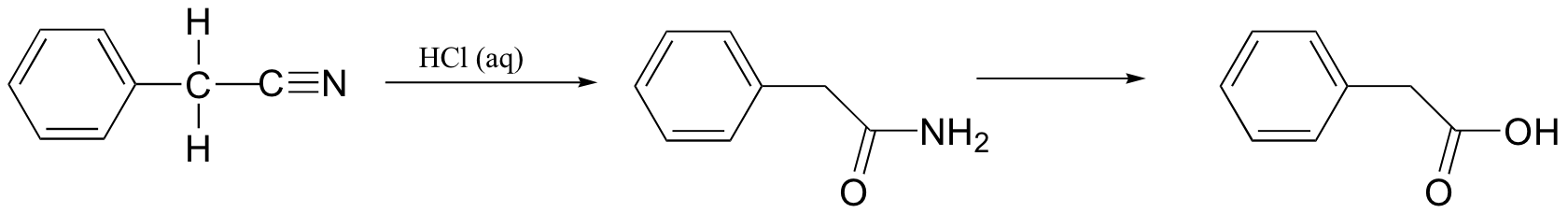
P12.18: In the lab, nitrile groups can be hydrolyzed by treatment with aqueous acid. In this reaction, the nitrile group is first converted to an amide, and then to a carboxylic acid. Draw a mechanism for this process.
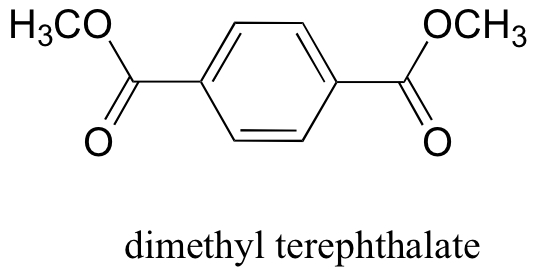
P12.19: The polyester fabric known as Mylar is formed from the polymerization of dimethyl terephthalate with ethylene glycol (CH2OHCH2OH) as a 'linker' molecule Propose a reasonable mechanism showing the beginning of this polymerization process: in other words, showing the formation of a mini-polymer containing two dimethyl terephthalate units linked by one ethylene glycol unit.
P12.20: Draw mechanisms for the conversion of acetic acid to acetic acid chloride (section 12.2D) using
a) SOCl2
b) PCl3
Challenge problems
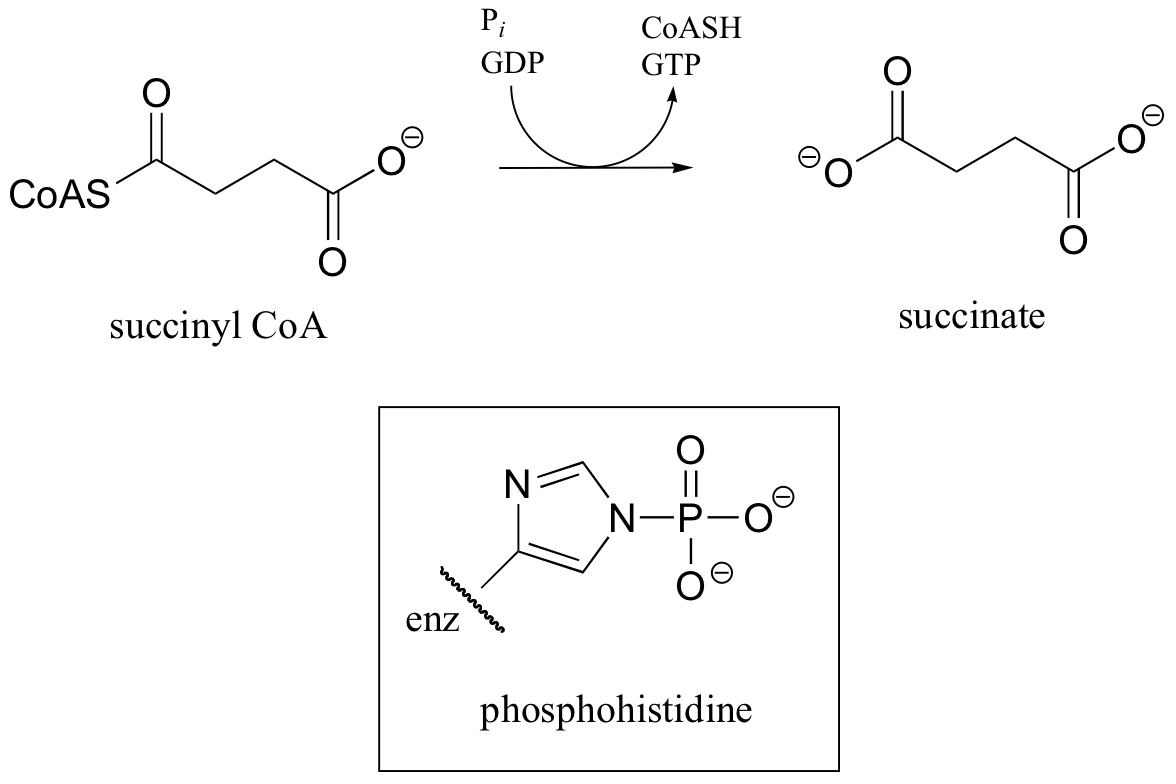
C12.1: In a step in the citric acid cycle, hydrolysis of succinyl-CoA is coupled to phosphorylation of GDP. The mechanism involves the transient phosphorylation of an active site histidine. Suggest a (multi-step) mechanism for this process.
C12.2: A 14C-labeled diazoketone compound (structure below) was used to irreversibly inactivate an enzyme called glutaminase A. Inactivation was shown to occur with 14C labeling of an active site cysteine.
a)Propose a mechanism of inactivation and cysteine labeling.
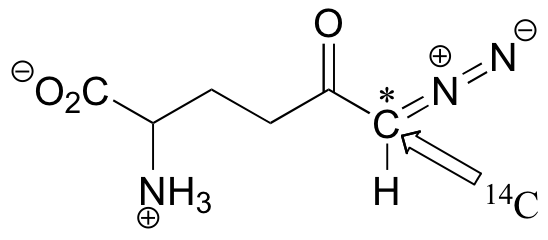
b) To a lesser extent, inactivation of the enzyme and labeling of the cysteine was found to occur with release of a radioactive compound from the active site. Propose a mechanism for the mode of inactivation.
C12.3: Dehelogenase enzymes catalyze the cleavage of carbon-halogen bonds, and are of interest to scientists looking for new ways to detoxify organohalogen pollutants that make their way into the environment. One such dehalogenase catalyzes the following reaction:
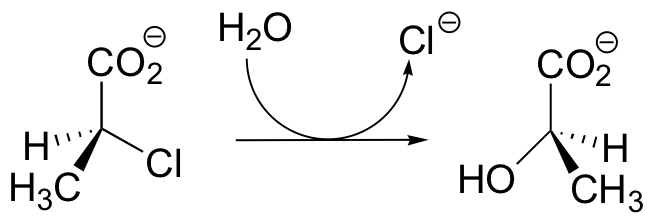
An active site aspartate is thought to carry out the initial nucleophilic attack that expels the chloride.
a) Draw a likely mechanism for the complete reaction shown above. Look carefully at the stereochemical progress!
b) When the active site aspartate was mutated to asparagine, the enzyme still maintained activity. Mass spectrometry analysis indicated that, at one point in the catalytic cycle of the mutant enzyme, the asparagine exists as a cyanoalanine. Draw a likely mechanism for the reaction as catalyzed by the mutant enzyme, including formation of the transient cyanoalanine residue and subsequent regeneration of the asparagine.
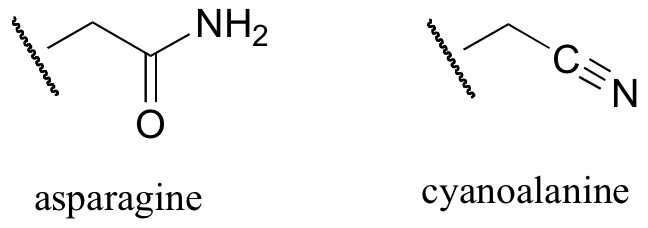
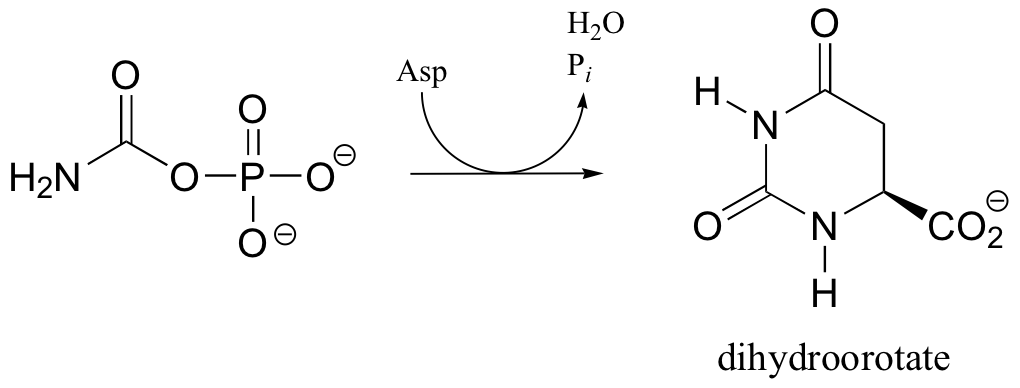
C12.4: In the biosynthetic pathway for the DNA/RNA bases uridine and cytidine, a reaction occurs in which carbamoyl phosphate condenses with aspartate, and the resulting intermediate cyclizes to form dihydroorotate. Propose a mechanism for this transformation. Hint: in a rather unusual step, a carboylate group is at one point in the process directly subjected to an acyl transfer reaction, without prior phosphorylation. The enzyme accomplishes this with the help of two bound zinc ions, which stabilize the negative charge on a hydroxide leaving group.