Extra Credit 27
- Page ID
- 83422
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Q19.9B
Calculate Ecell using the Nernst equation for the following cells.
- \(Zn_{(s)}|Zn^{2+}_{(aq)}(0.1M)||Sn^{2+}_{(aq)}(0.8M)|Sn_{(s)}\)
- Cu(s)|Cu+(aq)(0.4M)||F−(aq)(O.9M)|F2(g)(0.5atm)|Pt(s)
S19.9B
Step 1: Make half reactions and determine the oxidation and reduction equation. (Make sure equation is balanced!)
Step 2: Find the E° for each half reactions using the standard reduction potential table.
Step 3: Add the E° of both half reactions together to find the E°cell.
Step 4: Determine the number of moles of electrons transferred (n).
Step 5: Calculate the reaction quotient (Q). Q=[Products]/[Reactants]
Step 6: Calculate the Ecell (Plug and chug). Ecell = E°cell - (0.0592/n)logQ at 298K.
a) Zn(s)|Zn2+(aq)(0.1M)||Sn2+(aq)(.8M)|Sn(s)
Ox:Zn2+(aq) + 2e- → Zn(s) E° : -0.763
Red: Sn2+(aq) + 2e- → Sn(s) E° : -0.137
E°cell : -0.9
Phase II corrections in red: To find \(E^o_{cell}\), you should subtract \(E_{cathode}-E_{anode}\). This gives use \(E^o_{cell}\)= -.137-(-.763)= 0.626
n=2
Q=\([Zn^{2+}]/[Sn^{2+}]\)=0.10/0.8=0.125
Ecell = E°cell - 0.0592/n(log[Q]) = -0.9 - 0.0592/n(log(0.125))
Ecell = -0.8733V
Therefore, the answer should be 0.653 V if you use \(E^o_{cell}\)=0.626
b) Cu(s)|Cu+(aq)(0.4M)||F−(aq)(O.9M)|F2(g)(0.5atm)|Pt(s)
Anode: Cu+(aq) + e- → Cu(s) E° = 0.52
Cathode: F2(g) + 2e- → 2F-(aq) E° = 2.866
E°cell = 2.186
\(E^o_{cell}\) should be \(E_{cathode}-E_{anode}\)= 2.866-0.52=2.346 V
n=2
Q=[Cu2+][F−]=0.40.9=0.4444
Q should be based on the balanced chemical equation: Q=\([Cu^+]^2/[F^-]\)=0.178
Ecell = E°cell - 0.0592/nlog[Q]
= 2.186 - 0.0592/nlog(0.4444)
Ecell = 2.196V
\(E_{cell}\) should be 2.346-(0.0592/2)log(0.178)=2.368 V
Q19.72A
Consider the two following reduction half reactions:
Sn4++2e−→Sn2+
Sn2++2e−→Sn(s)
Calculate the E∘reduction for the reaction
Sn4++4e−→Sn(s)
S19.72A
We will need to combine these equations, but we can’t simply add the E∘cell values together. We will need to convert and find the ΔG values for each equation. First, find the E∘cell of the two given half reactions: they are 0.154 V, and -0.137 V respectively from Table P2.
The ΔG of the first equation
ΔG∘=nFE∘cell
Plugging in the values given will simplify to
=−2F(0.154)
ΔG∘=2F(0.154)
where F is Faraday's constant.
2nd equation =−2∗F∗(−0.137)
ΔG for the desired equation =(−0.308F)V+(0.274F)V
\(\Delta G^o\) values are additive.
ΔG=(−0.034F)V
Now to find the E∘cell=((−0.034F)/4F)∗V=−0.0085V=E∘cell
To find this, he simply solved the above Nernst Equation for \(E^o_{cell}\). Faraday's constant is 96.485 \(\frac{C}{mol}\)
It should, however, be a positive value: \(E^o_{cell}\) = 0.0085 V
Q21.5D
Draw Lewis structures for the following ligands:
- NH3
- en
- SCN−
- NO2-
S21.5D
Step 1: Find the total number of valence electrons.
Step 2: Elements up to period 4 generally follow the "Octet Rule," meaning they need 8 electrons to fill their outer shell.
Step 3: Determine the central atom. It is usually the atom with the highest valence or the least electronegative.
Step 4: Place electrons on the outside atoms first, the remaining electrons go on the central atom (creating lone pairs and/or double bonds, triple bonds, etc).
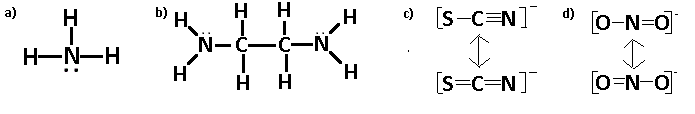
It would be useful to mention the fact that these lines you see are shared bonds between atoms, and they count for two of the total number of valence electrons. Also, in the case of (b), there is not really a central atom, as you can see by the symmetric nature of the molecule. The two dots on the nitrogen are a lone pair, and they each count as one electron, for a total of two per pair. Parts (c) and (d) have those arrows to indicate a resonance structure, because each molecule that has different types (single vs. double, etc.) of bonds may have all of them manifested, so we must draw all possible resonance structures. Lastly, which should be firstly, you must calculate the valence electrons based on the group that each atom is in- Carbon is in group 4A, so it has 4 valence electrons, oxygen is in group 6A so it has 6 valence electrons, etc. If the molecule has a charge, there are that many more total valence electrons (e.g. 2- has 2 more, 2+ has two less).
Q24.13D
Use the date table below to determine the rate law of the reaction: A+B→2D
| Experiment | Initial [A],M | Initial [B],M | Initial Rate of Reaction, Ms-1 |
| 1 | 1.5 | 1.5 | 4.3 × 10-3 |
| 2 | 1.5 | 3.0 | 8.6 × 10-3 |
| 3 | 3.0 | 3.0 | 1.12 × 10-2 |
S24.13D
For more help see: The Rate of a Chemical Reaction
From looking at the table, [A] is first order and [B] is first order
overall rxn is 2nd order
R=k[A][B]
4.3E-3Ms−1=k(1.5M)(1.5M)
k=1.9E-3(M−1)(s−1)
k=\(\frac{1.9*10^{-3}}{M*s}\)
The solution is correct. To add to this, it is useful to say that we look at how increasing the concentration of one reactant affects the rate of reaction. In this case, when we double [A], reaction rate doubles. This is a 1:1 increase, so we call it first order. It is the same for [B].
Q24.61A
Please answer the following about the catalytic activity of both platinum metal and enzymes:
- Where are there active sites?
- Are they heterogeneous or homogeneous?
- Are they specific or nonspecific?
S24.61A
- The active site of platinum and of enzymes are at a metal center.
- Enzymes are usually homogeneous, meaning they are soluble in the reactant; platinum, however, is heterogeneous, meaning it cannot be dissolved in the reactant.
- Enzymes are extremely specific to their substrates, while platinum is usually more nonspecific.
The active site of an enzyme is where it binds to its substrate.
Q25.29C
A science fiction writer is writing a short story about a team of scientists landing on Jupiter's moon for the first time. In their exploration, they find a large obelisk with some kind of electromagnetic field powered by an ancient fission reactor. Since the scientists' space suits already protect against the harmful radiation of interplanetary space, they decide to collect a sample and use it to find an approximate absolute age for the ship. The scientists determine that the reactor was able to run efficiently using protactinium-231 as fuel (remember that this is, indeed, fiction). If the scientists report to mission control that the reactor has been running for 30,000 years, what ratio of protactinium-231 to Uranium-235 and its derivatives was observed in the fictional field laboratory?
S25.29C
The half life of Protactinium-231 is 32,760 years. We use λ=0.693/t1/2 to determine the rate constant of the reaction.
λ=0.693/32,760y=2.11×10E−5y−1
\(\lambda=\frac{.693}{t_{1/2}}\) = \(\frac{.693}{32760 years}\) = 2.12*\(10^{-5}y^{-1}\)
Now, using the equation: ln(Nt/N0)=−λt, we can determine the ratio of moles of the protactinium-231 to Uranium-235 (and its derivatives)
ln(Nt/N0)=−λt⇒(Nt/N0)=e^(−λt)⇒(Nt/N0)=e^[−(2.11×10E-005)(30,000)]=0.53
Knowing that the molar ratio of Protactinium-231 to Uranium-235 (and derivatives) is 0.53, we can determine the mass ratio of the sample collected fictional scientists.
Since there is a ratio of 0.53, that means there is 0.53 protactinium-231 moles per 1 mole of sample and conversely that there are 0.47 moles of Uranium-235 and its derivatives for every 1.00 mole of sample.
So, using some conversion factors for the molar mass of the species, we can determine that the ratio of Protactinium-231 to Uranium-235 was:
[0.47 moles U-235/0.53 moles Pa-231]×[235 g of U-235/1 mole of U-235]×[1 mole of Pa-231/231 g Pa-231]=110.45 g U-235/122.43 g Pa-231 -->
1 g U-235/1.108 g Pa-231.
\(\frac{0.47 mol U-235}{0.53 mol Pa-231}*\frac{235 g U-235}{1 mol U-235}*\frac{1 mol Pa-231}{231 g Pa-231}\)=\(\frac{1 g U-235}{1.108 g Pa-231}\)
If the fictional Scientists reported that the artifact they found was 30,000 years old, they would have found 1 g of U-235 and its derivatives for every 1.108 g of Pa-231 fuel in the reactor.
Q21.1.6
List the three primary sources of naturally occurring radiation. Explain the factors that influence the dose that one receives throughout the year. Which is the largest contributor to overall exposure? Which is the most hazardous?
S21.1.6
Three primary naturally occurring radiations are radium, uranium and thorium, each all having long half lives. Inhalation of air, ingestion of food and water, terrestrial radiation from the ground and cosmic radiation from space are all factors that influence the doses that a person receives throughout the year. Inhalation of the air is the largest contributor to exposure. Radiation can damage DNA or kill cells. When radiation is exposed to your body, it will collide with atoms and this will change and damage your DNA.
According to the LibreTexts page on Ionizing Radiation, some other factors that can influence the dose of radiation that a person receives include their place of residence (those in Denver experience more radiation due to cosmic rays than those at sea level), habits such as smoking, and even the materials a building is made out of.
Q24.6.3
Will the value of Δo increase or decrease if I− ligands are replaced by NO2− ligands? Why?
S24.6.3
The value of Δo would increase because NO2− is a stronger ligand which means that it would have a larger split.
Yes, strong-field inducing ligands increase the crystal field splitting of the molecule, meaning that the electrons have to absorb a shorter wavelength of light to become excited.

