12.6: Translation
- Page ID
- 165340
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The Mechanism of Protein Synthesis (Translation)
Just as with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination. The process of translation is similar in bacteria, archaea and eukaryotes.
Translation Initiation
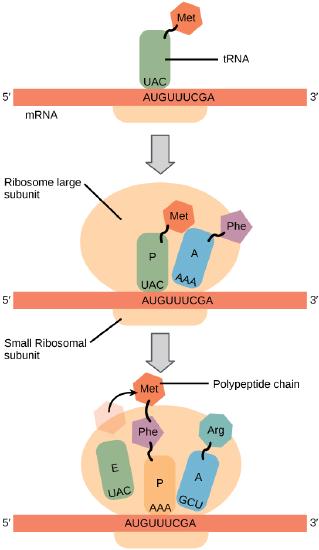
In general, protein synthesis begins with the formation of an initiation complex. The small subunit binds to a site upstream (on the 5' side) of the start of the mRNA. It proceeds to scan the mRNA in the 5'-->3' direction until it encounters the START codon (AUG). The large subunit attaches and the initiator tRNA, which carries methionine (Met), binds to the P site on the ribosome.
The small ribosomal subunit will bind to the mRNA at the ribosomal binding site. Soon after, the methionine-tRNA will bind to the AUG start codon (through complementary binding with its anticodon). This complex is then joined by large ribosomal subunit. This initiation complex then recruits the second tRNA and thus translation begins.The small subunit It proceeds to scan the mRNA in the 5'-->3' direction until it encounters the START codon (AUG). The large subunit attaches and the initiator tRNA, which carries methionine (Met), binds to the P site on the ribosome.
Bacterial vs Eukaryotic initiation
Protein synthesis begins at an AUG (met) codon- but proteins may have many methionines, and mRNAs may have many AUGs. How does the ribosome know where to begin?
In prokayrotic mRNA, a sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (AGGAGG), base-pairs with a rRNA molecule within the small subunit of bacterial and archeal ribosomes. This interaction anchors the 30S ribosomal subunit at a precise location on the mRNA template.
Instead of binding at the Shine-Dalgarno sequence, the eukaryotic initiation complex (a number of proteins in addition to the small subunit) recognizes the 7-methylguanosine cap at the 5' end of the mRNA. Once at the cap, the initiation complex tracks along the mRNA in the 5' to 3' direction, searching for the AUG start codon. Many eukaryotic mRNAs are translated from the first AUG, but this is not always the case. The nucleotides around the AUG affect the probability that it will be chosen as the start codon, and the consensus sequence varies between species. The "helper" proteins of the initiation complex fall off once the large subunit is loaded.
Note that in both cases the selection of an AUG establishes the reading frame (one of a possible 3) for the entire protein. A very important difference between these modes of start-site selection is that a single prokaryotic transcript can potentially encode several sequential proteins, as the ribosome can scan the entire length of the message for Shine-Dagarno sequences. Often the several proteins involved in a single process (subunits of a holoenzyme. or sequential steps in a metabolic pathway) are encoded on a single message. In contrast, in eukaryotic nuclear genes, each transcript only encodes a single protein (as always, there are exceptions).
Translation Elongation
During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into 'view', and specific binding with the corresponding charged tRNA anticodon is ensured. If mRNA were not present in the elongation complex, the ribosome would bind tRNAs nonspecifically.
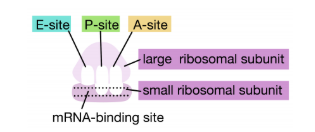
The large ribosomal subunit consists of three compartments: the A site binds incoming charged tRNAs (tRNAs with their attached specific amino acids), the P site binds charged tRNAs carrying amino acids that have formed bonds with the growing polypeptide chain but have not yet dissociated from their corresponding tRNA, and the E site which releases dissociated tRNAs so they can be recharged with another free amino acid.
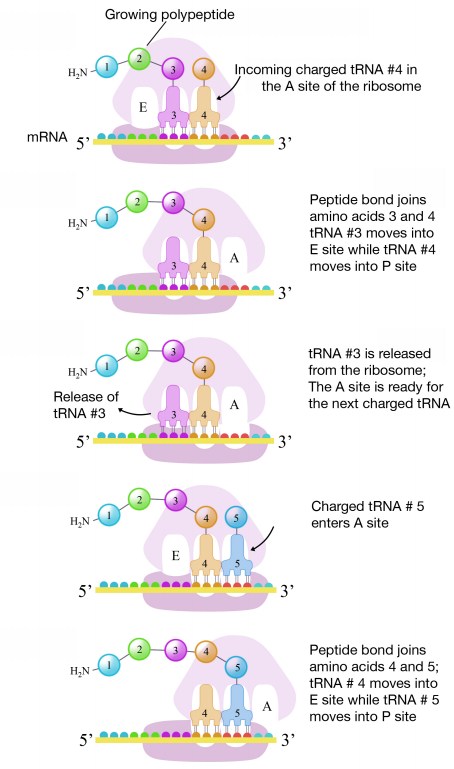
A tRNA bound to its amino acid (known as an aminoacyl-tRNA) that is able to base pair with the next codon on the mRNA arrives at the A site. The preceding amino acid (Met at the start of translation) is covalently linked to the incoming amino acid with a peptide bond. The initiator tRNA moves to the E site and the ribosome moves one codon downstream. This shifts the more most recent tRNA from the A site to the P site, opening up the A site for the arrival of a new aminoacyl-tRNA. Polypeptide synthesis repeats, the tRNA residing in the E site is released from the complex, the tRNAs in the P site and A site shift over and the next amino acid is added to the growing polypeptide chain. This cycle repeats until a stop codon is reached.
Ribosomal steps are induced by conformational changes that advance the ribosome by three bases in the 3' direction. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP. Peptide bonds form between the amino group of the amino acid attached to the A-site tRNA and the carboxyl group of the amino acid attached to the P-site tRNA. The formation of each peptide bond is catalyzed by peptidyl transferase, a catalytic RNA (surprise! not a protein) that is integrated into the 50S ribosomal subunit. The energy for each peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor. The amino acid bound to the P-site tRNA is linked to the growing polypeptide chain. As the ribosome steps across the mRNA, the former P-site tRNA enters the E site, detaches from the amino acid, and is expelled (it will be recharged by tRNA synthetase later). The ribosome moves along the mRNA, one codon at a time, catalyzing each process that occurs in the three sites. With each step, a charged tRNA enters the complex, the polypeptide becomes one amino acid longer, and an uncharged tRNA departs.
This and subsequent steps in the synthesis of the polypeptide are called the elongation phase of translation. Once the first two amino acids are linked , the first tRNA dissociates, and moves out of the P-site and into the E, or Exit site. The second tRNA then moves into the P-site, vacating the A-site for the tRNA corresponding to the next codon.
Termination
Translation ends when the ribosome reaches a STOP codon (UAA, UAG or UGA). There are no tRNA molecules with anticodons for stop codons, instead protein release factors recognize these codons when they arrive at the A site. Binding of a release protein causes the polypeptide (protein) to be released from the ribosome. The ribosome subunits dissociate (split) from each other and can be reassembled later for another round of protein synthesis.
Post-translational Protein Modification
After translation individual amino acids may be chemically modified. These modifications add chemical variation not present in the genetically encoded amino acids, and new properties that are rooted in the chemistries of the functional groups that are being added. Common modifications include phosphate groups, methyl, acetate, and amide groups. Some proteins, typically targeted to membranes, will be lipidated - a lipid will be added. Other proteins will be glycosylated - a sugar will be added. Another common post-translational modification is cleavage or linking of parts of the protein itself. Signal-peptides may be cleaved, parts may be excised from the middle of the protein, or new covalent linkages may be made between cysteine or other amino acid side chains. Nearly all modifications will be catalyzed by enzymes and all change the functional behavior of the protein.
Summary
An mRNA is used to synthesize proteins by the process of translation. The genetic code is the correspondence between the three-nucleotide mRNA codon and an amino acid. The genetic code is “translated” by the tRNA molecules, which associate a specific codon with a specific amino acid. The genetic code is degenerate because 64 triplet codons in mRNA specify only 20 amino acids and three stop codons. This means that more than one codon corresponds to an amino acid. Almost every species on the planet uses the same genetic code; the "deviant codes" are not radically different, but change the meaning of one or two codons. More impressive exceptions are species that encode 21 or 22 amino acids, rather than the usual 20.
The players in translation include the mRNA template, ribosomes, tRNAs, and various enzymatic factors. The small ribosomal subunit binds to the mRNA template. Translation begins at the initiating AUG on the mRNA (this also establishes the reading frame). The formation of bonds occurs between sequential amino acids specified by the mRNA template according to the genetic code. The ribosome accepts charged tRNAs, and as it steps along the mRNA, it catalyzes bonding between the new amino acid and the end of the growing polypeptide. The entire mRNA is translated in three-nucleotide “steps” of the ribosome. When a stop codon is encountered, a release factor binds and dissociates the components and frees the new protein.


