3.4.3. Ion Exchange Chromatography
- Page ID
- 165275
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In ion exchange chromatography, the support consists of tiny beads to which are attached chemicals possessing a charge. Each charged molecule has a counter-ion. The figure shows the beads (blue) with negatively charged groups (red) attached. In this example, the counter-ion is sodium, which is positively charged. The negatively charged groups are unable to leave the beads, due to their covalent attachment, but the counter- ions can be “exchanged" for molecules of the same charge. Thus, in cation exchange column, the chemical groups attached to the beads are negatively charged groups and will have positively charged counter-ions and positively charged compounds present in a mixture passed through the column will exchange with the counter-ions and “stick" to the negatively charged groups on the beads. Molecules in the sample that are neutral or negatively charged will pass quickly through the column. On the other hand, in anion exchange chromatography, the chemical groups attached to the beads are positively charged and the counter-ions are negatively charged. Molecules in the sample that are negatively charged will “stick" and other molecules will pass through quickly. To remove the molecules “stuck" to a column, one simply needs to add a high concentration of the appropriate counter-ions to displace and release them. This method allows the recovery of all components of the mixture that share the same charge.
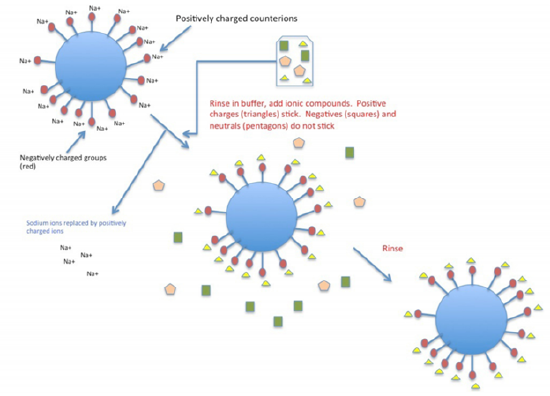
Figure 3.4.3.1: Cation exchange chromatography
Usually, samples are loaded under low ionic strength conditions and bound material is eluted using either a step or gradient elution of buffer with higher ionic strength.
Generally speaking, a protein will bind to a cation exchange resin if the buffer pH is lower than the isoelectric point (pI) of the protein, and will bind to an anion exchange resin if the pH is higher than the pI.
.png?revision=1&size=bestfit&width=497&height=416)
Knowledge of the pI of the protein is therefore helpful in designing a purification protocol using ion exchange resins (however, you can always simply try different resins to see which works best).
Elution of proteins from ion exchange resins
- Proteins bound to ion exchange resins are bound via non-covalent ionic (salt-bridge) interactions. We can compete for these ionic binding sites on the resin with other ionic groups, namely, salts
- There are two general types of methods when eluting with a salt solution: 1. Gradient elution and 2. Step elution
- A gradient elution refers to a smooth transition of salt concentration (from low to high) in the elution buffer. Weakly binding proteins elute first, and stronger binding proteins elute last (i.e. they require higher salt concentrations in the buffer to compete them off the column)
- A gradient salt concentration can be made using a gradient maker. In its simplest form, this consists of two containers (must be the same shape) connected by a siphon (or tube at the bottom). One container contains the low salt buffer, and the other contains high salt buffer. The buffer is withdrawn from the low salt container:
.png?revision=1&size=bestfit&width=396&height=344)
Figure 3.4.3.2: Gradient maker
This will produce a linear gradient from low to high salt concentrations over the total volume of the gradient
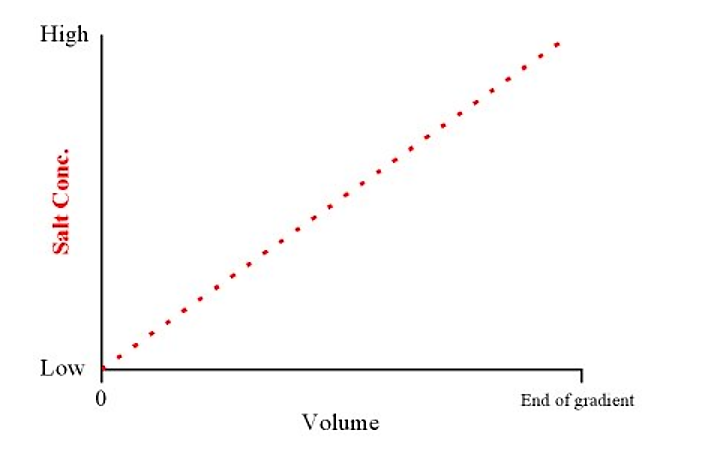.png?revision=1)
Figure 3.4.3.3: Salt concentration and volume
If we know the concentration range of salt over which a protein of interest will elute we can simply elute with a buffer containing that concentration of salt. This is known as a step elution.
Step elutions are generally faster to run, and elute the protein in a smaller overall volume than with gradient elutions. They generally work best when contaminants elute at a significantly different salt concentration than the protein of interest.
.png?revision=1&size=bestfit&width=428&height=286)
Figure 3.4.3.4: Step elution
Note that after ion exchange chromatography the protein of interest will be in a buffer with a potentially high salt concentration. This must be taken into account before proceeding with the next step in the purification scheme.
Dialysis
- After an ammonium sulfate precipitation step, or an ion exchange chromatography step, the protein of interest may be in a high salt buffer. This may be undesirable for several reasons. How do we get rid of salt in our sample?
- One of the most common methods is that of dialysis
- The method of dialysis makes use of semi-permeable membranes. In the simplest example, this membrane is manufactured in the form of tubing (looking much like a sausage casing)
- The main feature of this membrane is that it is porous. However, the pore size is such that while small salt ions can freely pass through the membrane, larger protein molecules cannot (i.e. they are retained). Thus, dialysis membranes are characterized by the molecular mass of the smallest typical globular protein which it will retain.
- This is commonly referred to as the cutoff of the tubing (e.g. Spectrapore #6 dialysis tubing has a cutoff of 1,000 Daltons, meaning that a 1,000 Dalton protein will be retained by the tubing but that smaller molecular mass solutes will pass through the tubing)
- Dialysis proceeds by placing a high salt sample in dialysis tubing (i.e. the dialysis "bag") and putting it into the desired low salt buffer.
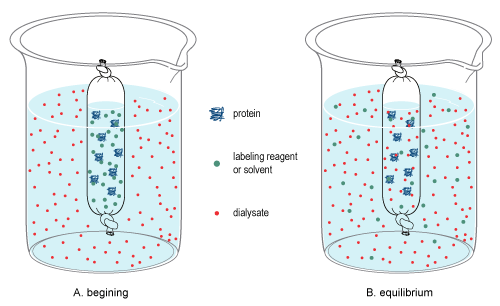
Figure 3.4.3.5: Dialysis (https://en.wikipedia.org/wiki/Dialysis_(biochemistry)
Over time the concentration of low molecular mass solutes within the bag, and in the low salt buffer, will come achieve equilibrium. In practical terms (for the above case) salt molecules will diffuse out of the bag into the low salt buffer.
.png?revision=1)
Figure 3.4.3.6: Salt diffusion
Concentration
- What if our protein sample is too dilute for our needs? How can we concentrate our samples?
- One common method is, again, to use a semi-permeable membrane for this purpose.
- A very simple method is to place our sample in a dialysis bag and coat it with a high molecular weight solute which can readily be dissolved by the buffer.
- In another variation, the semi-permeable membrane is manufactured into a flat disk and placed at the bottom of a container which holds our sample. In one method the container is pressurized and forces buffer out of the container (protein is retained and is concentrated). In another method, the vessel is centrifuged and the centripetal force achieves the same goal as pressure in the previous example.
For both dialysis and concentration, it is essential that the membrane does not interact with the protein (i.e. has no affinity for, and will not bind, the protein)
Contributors
Dr. Kevin Ahern and Dr. Indira Rajagopal (Oregon State University)


