Signaling: Calcium and Calmodulin
- Page ID
- 67247
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Calmodulin
This is a video introduction to the calcium signaling protein, calmodulin, from an inorganic chemist's perspective!
Calmodulin, or CaM, is a polypeptide that is ubiquitous in all eukaryotic cells. This protein is known as calmodulin because it is a calcium-modulated protein that plays a vital role in the process of calcium signal transduction. Calcium signal transduction is the process through which the interactions between calcium ions and numerous proteins mediate communication between cells. calmodulin’s function, therefore, is necessary in all eukaryotic cells, and some of the tasks that it helps to accomplish are nerve signaling, skeletal muscle movement, and memory. By sensing calcium ions in the environment, calmodulin activates and subsequently acts as an intermediate, initiating the binding of important proteins such as kinases, assisting our cells in basic and sophisticated function (“Calmodulin”).
The video below demonstrates the structural change that occurs within CaM during calcium binding (PDB codes 3CLN and 1CFD).
The protein itself is 148 amino acids in length with two globular regions containing 2 EF-hand motifs each, which are characteristic sites of calcium-mediated polypeptides. When activated, calmodulin houses 4 Ca2+ ions that drastically change the shape of the protein. When calmodulin binds with the calcium ions, the protein opens from its apo form to its halo form, exposing an alpha helix that is known as the linker or central tether region. Coined for its flexibility, the central tether region is the location of the protein on which partner proteins bind and contribute to the cascade that is the secondary messaging of calcium. Based on its structure and its need for calcium ions to function, calmodulin must be able to select for calcium ions in the cytoplasm, and the interactions between the ion and the ligands in the EF hand domains support this idea of selectivity (Bertini et. al., 635-638).
This image is a depiction of how an EF-hand motif resembles a hand. The ribbon diagram on the left shows one of the four characteristic helix-turn-helix EF hand motifs of calmodulin. (PDB code 3CLN)
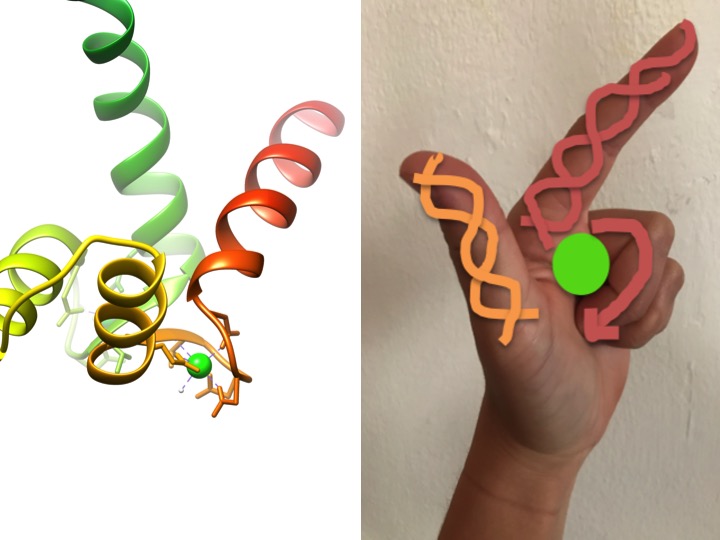
EF-hand motifs are highly conserved structural regions of proteins involved in the binding of calcium. These regions are known as “hand” motifs because they resemble a hand that is made in the shape of an L, where the metal ion is located in the middle of the fist, the alpha helices are indicated by the upward-pointing index finger and inward-facing thumb, and the curled fingers represent the turn or loop region. This is demonstrated in the figure above. In a characteristic EF-hand motif, amino acids including glutamates, asparagines, aspartic acids, and glutamic acids bind to Ca, as well as water (Bertini et. al. 639). The typical EF-hand domain bonding sequence is shown in the figure below.
This cartoon illustrates the coordinated amino acids in a typical EF hand domain (The exact amino acid composition varies!). The dashed line represents coordination of Ca to the oxygen of a backbone carbonyl, while solid lines indicate coordination to side chains or water.
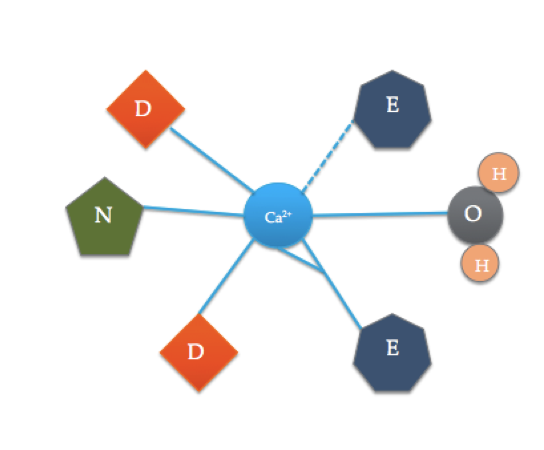
The calmodulin binding site is somewhat different than the most typical EF hand domain shown above. The 6 ligands of calmodulin include the side chains of three asparatic acids (D), 1 glutamic acid (E) that forms two coordinate covalent bonds with the ion, 1 water molecule from solution, and 1 carbonyl molecule from the backbone. The binding site of calmodulin consist of a Ca2+ ion surrounded by 6 ligands while forming 7 bonds.
One can notice that the EF motif within calmodulin includes many of the same ligands, just in different places. This conformation may lend an idea of why calmodulin is selective for calcium and how only the presence of a calcium ion can satisfy the binding site that causes the protein to activate. This is related to the thermodynamics of calmodulin within the cell and how it responds to the presence of calcium ions.
To understand the thermodynamics that couples the activation of calmodulin, one must first understand the concentration of ions such as calcium and magnesium within a eukaryotic cell, including that of a human. The concentration of these ions must be closely regulated. Within in the cell, calcium signaling is accompanied by a temporary increase in the concentration of calcium ions, which is sensed by proteins such as calmodulin (Bertini et. al., 636). This calcium surge could be due to intracellular G-proteins that induce the rough and smooth reticulum to release calcium, or the calcium ions could be brought in from the extracellular space. In the case of calmodulin, it is usually responding to calcium being brought into the cell from the outside, which occurs during processes such as nerve signaling (Bertini et. al. 637). Before the concentration of calcium is momentarily raised, the concentration within the cell is usually between 10-100 nm, whereas during the brief influx of ions, the concentration increases to 1,000-100,000 nm. The change in concentration causes the calmodulin to sense the calcium ions, bind them, and initiate further signal transduction (Bertini et. al., 635).
How, then, does calmodulin definitively bind calcium ions and not, for example, magnesium ions? Mg2+ ions are actually quite similar to calcium ions: they would engage in electrostatic interactions rather than covalent, and they are considered hard atoms. The reason that calmodulin seems to favor calcium ions rather than magnesium ions is related to the binding constants of each ion. Calmodulin can exist while bound to other ions, such as magnesium, but if there is a higher concentration of calcium, the calcium ions will immediately compete out the magnesium ions to occupy calcium-binding sites. This is because the binding constant of CaM and Ca2+ (Ka) is larger than that of CaM and Mg2+, as shown in Figure 7. This indicates that when bound to calcium ions, calmodulin would produce more products. One must consider all of these ideas keeping in mind that this process happens innumerable times per second, and the turnover of this protein changes to respond to the ever-modulated calcium concentration of the cell.
Why is calmodulin binding with calcium ions more favorable than with other metals? It could be because of the size of the ions in relation to the steric hindrance induced by the ligands. The binding site is characterized by 6 ligands making 7 coordinate bonds. Because calcium ions are larger than magnesium ions, as demonstrated in Figure 8, it is possible that calcium is favored because it increases the distance of the ligands from each other, therefore decreasing steric interactions between the ligands. The very structure of the protein itself could also pose a reason for the selectivity of calcium ions. Overall, it is known that due to thermodynamics based on varying cellular concentration of calcium ions, calmodulin will sense the increase of ions, bind them, and help to carry out its function.
CaM + Ca2+ <--> Ca2+CaM (KCaM-Ca)
CaM + Mg2+ <--> Mg2+CaM (KCaM-Mg)
Mg2+-CaM + Ca2+ <--> Ca2+CaM + Mg2+
because KCaM-Ca > KCaM-Mg
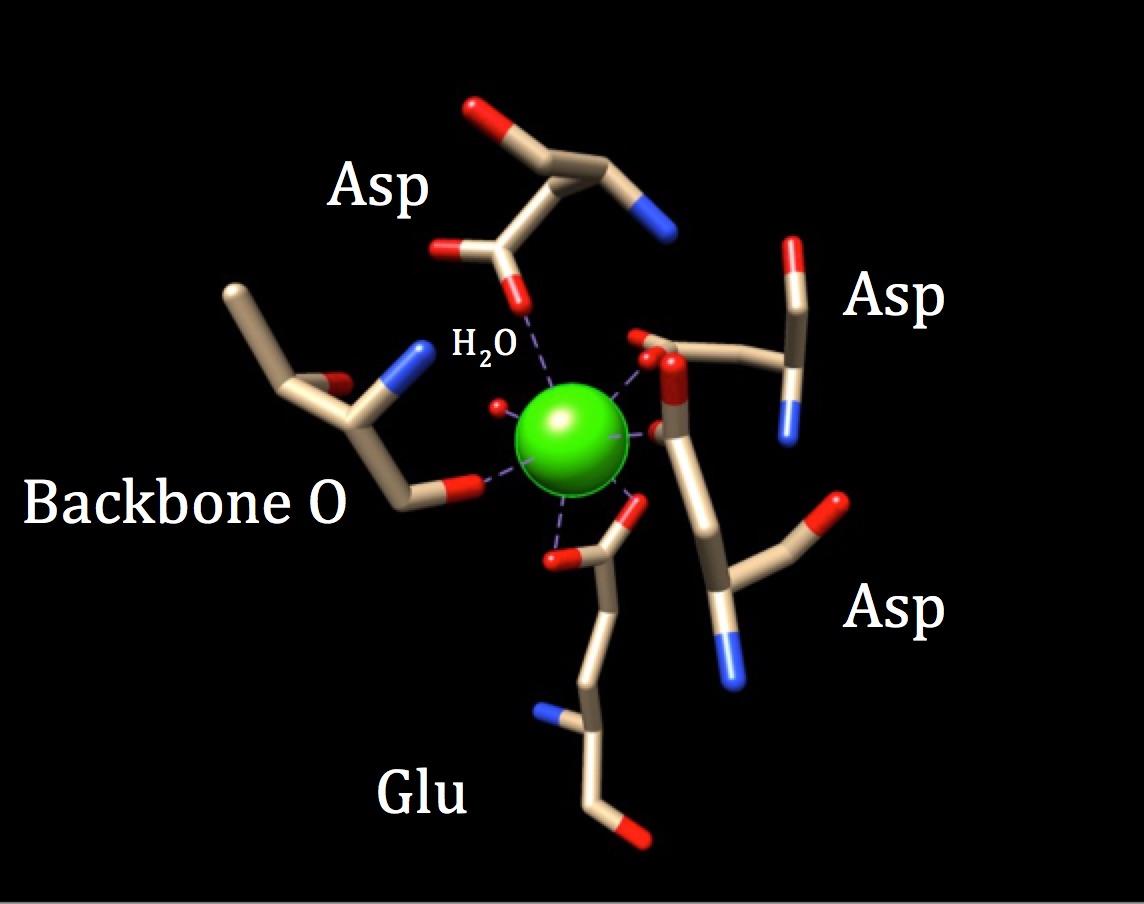
In a further discussion of the chemistry of the binding site of calmodulin, the coordination geometry should be explored. Because the calcium ion interacts with seven donor atoms, the system is known as a heptadentate system. Because calmodulin is surrounded by six ligands, one would assume an octahedral coordination geometry. But, because one of the ligands interacts in a bidentate fashion with the ion—meaning that it has two coordinate covalent bonds coming from different atoms in one amino acid—the coordination complex takes on a distorted octahedral shape. The specific coordination is outlined in the figure below.
The coordination geometry of calmodulin's calcium-binding site. The center calcium metal ion is surrounded by three asparagines (Asp), one backbone oxygen, one gluatmic acid (Glu) and one water molecule. (PDB code 3CLN)
We can also refer to the theory of the chelate effect to better understand the ability of calmodulin to bind to metals ions with high affinity. A chelator is an atom that is able to bind with more than one donor atom. Polydentate bonding is related to chelate effect, which states that when a ligand binds with many donor groups (poly), the coordinate covalent bonds are much stronger, or more tightly held. This theory is backed up by thermodynamics, specifically that polydentate binding increase the entropy of a system, making it more positive (more disorder), which is thermodynamically favorable. Applying this theory to calmodulin helps to explain not only the importance of the polydendate bonding within the sites themselves, but also the need for the protein to bind 4 different calcium ions to activate its function. By essentially requiring the bonding of four Ca2+ ions within the sites, the system encourages an increase in entropy by introducing more substrates/reactants to form a greater number of products. This occurs due to the higher number of bonding within the ligand interactions of each bonding site. All in all, the binding of the calcium ion in the sites of the polypeptide are supported by the chelate effect theory and shed light on calmodulin’s efficiency in binding calcium.
EF-hand motifs have a certain structure based on the fact that calcium is the central metal. This is also true of the calmodulin-binding sites. The Ca2+ ion binds with like atoms in the structures of certain amino acids based on a theory called Hard/Soft Acid-Base Theory. This theory separates metals and ligands into two categories based on the polarizability and their charge to radius ratio. Hard acids and bases are smaller ions that have a larger charge to size ratio, and this leads them to interact in electrostatic ways (meaning attractions between + and – charges), whereas soft acids and bases are larger in size and therefore have a smaller charge to radius ratio, leading them to make bonds with covalent character. The phrase “like binds with like” refers to the fact that hard acids and bases tend to interact with other hard atoms, and the same goes for soft acids and bases. Ca2+ is characterized as a hard acid because, although it has a large radius, it interacts with ligands in a mostly electrostatic fashion. When looking at the calcium-binding site of calmodulin, one can see that the calcium ion binds to oxygen atoms of the 3 aspartic acids, glutamic acid, water, and backbone carbonyl. This binding is in line with “like binds with like” because water and the amino acids ligands, glutamic acid and asparatic acid, bind through oxygen donors which are characterized as hard bases. Figure 10 revisits the binding sites of calmodulin while also showing the negative sidechains of the amino acids that would interact with the calcium ions denoted in blue.
Concerning the donor atoms of the bound ligands and their ability to donate electron pairs to the coordination complex, the spectrochemical series explains how the atoms interact with the central metal. In the case of the calcium-binding site, the oxygens that interact with the Ca2+ ion are all deemed σ donors and π donors because the donor oxygens possess 2 or more pairs of electrons. The categories of the spectrochemical seires that specify electron availability and donation have implications on spin designation (low or high), which designates how d electrons fill in their orbitals, and delta (Δ) size between the metal’s d orbitals, but this does not make a difference for calcium because calcium’s d orbitals are unoccupied due to an absence of valence d electrons. This makes the d orbitals essentially negligible in most conversations concerning the protein. One exception is related to considering the lability of the calcium ion in the system.
The lack of electrons in Ca’s d orbitals yields zero ligand field stabilization energy, and explains the high lability of the calcium ion. Lability refers to the rate at which ligands can be replaced in coordination complexes, and therefore describes the kinetics of a system. Atoms are labile if they exchange ligands quickly. Because of its pertinent function in all eukaryotic cells, calmodulin is expected to bind and release calcium ions millions of times per second in order to send and receive messages from surrounding cells through the transfer of calcium ions. When regarding calmodulin’s ability to bind and release Ca2+ quickly, it is important to consider the occupancy of valence d electron orbitals. As stated, calcium possesses empty valence d orbitals. These unoccupied orbitals can give rise to an explanation for a Ca2+ ion’s lability. Because all of the calcium ion’s d orbitals are empty, it possesses a ligand field stabilization energy (LFSE) of 0. This indicates that the calcium is unstable when considering LFSE due to the fact that stable molecules usually have a negative value for LFSE. Instability in this case translates to a willingness to react quickly and move on and off complexes, supporting the claim that Ca2+ ions encourage fast reactions and can be characterized as labile. Though this seems to be counterintuitive to the discussion involving chelate effect, it is understood that these two theories do not contradict each other. The stability of the tightly held coordination bonds that is suggested by the chelate effect encourages the calmodulin to remain stable long enough to bind supplement proteins. Once that is completed and the concentration of calcium returns to normal, it is efficient in stripping the site of the calcium ions.
Calmodulin, based on its structure and the composition of its vital bonding site, plays an integral part of innumerable processes carried out by eukaryotic cells. Through the accepting and binding of calcium ions in signal transduction, calmodulin acts as a pivotal component of basic and high-level functioning in organisms such as humans. By applying bioinorganic theory and grasping an understanding of the delicately monitored environment of a eukaryotic cell, one can properly comprehend the ability of calmodulin to select for calcium ions and encourage further functioning involving more than 100 partner proteins.
Sources:
Bertini, Gray, Steifel, Valentine. Biological Inorganic Chemistry, Fifth Edition, W.H. Freeman and Co., New York, 2010.
“Calmodulin.” Wikipedia: The Free Encyclopedia. Wikimedia Foundation, Inc. 29 July 2016. Web. 7 September 2016.
Kuboniwa, H., N. Tjandra, S. Grzesiek, H. Ren, C. B. Klee, and A. Bax. “Solution Structure of Calcium-Free Calmodulin.” Nature Structural Biology 2, no. 9 (September 1995): 768–76. (PDB code 1CFD)
Babu, Y.Sudhakar, Charles E. Bugg, and William J. Cook. “Structure of Calmodulin Refined at 2.2 Å Resolution.” Journal of Molecular Biology 204, no. 1 (November 5, 1988): 191–204. doi:10.1016/0022-2836(88)90608-0. (PDB code 3CLN)
Contributed by:
This work was originally written by Makenzie Duncan, Fall 2016: Makenzie is currently (as of 2016) a junior biology major at Saint Mary's College in Notre Dame, IN.
This work was originally edited by Dr. Kathryn Haas, Assistant Professor at Saint Mary's College.

