Manganese Superoxide Dismutase (MnSOD)
- Page ID
- 98117
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Superoxide Anions
The reduction of O2 is an energy-producing process in cellular respiration or photosynthesis and is essential to aerobic life. The reduction occurs as a single-electron step that produces an oxygen radical, called the superoxide anion (O2ׁׄ͘͘•-), seen in Reaction 1. The free radicals produced are known as reactive oxygen species (ROS) and can cause cellular damage by oxidative stress, disruptions in bodily functions, and many diseases.3
(1)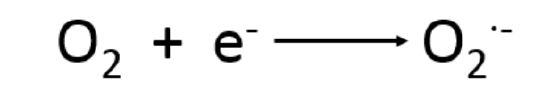
Reduction of oxygen, producing ROS. This reaction occurs in many processes in the body.
Superoxide Dismutases
Transition metal ions can adopt multiple oxidation states under in vivo condition. This ability allows transition metals to accept and donate electrons to carry out reduction and oxidation, or redox chemistry, utilized in numerous biological reactions. A redox catalyst is an enzyme that catalyzes biological redox reactions. All SODs catalyze the dismutation of O2˙ー radicals by protonating the coordination anion to produce H2O2 and O2 to prevent accumulation, leading to toxicity.1,3 Dismutation, or disproportionation, is when a reactant in a chemical reaction is simultaneously oxidized and reduced, producing two different substances. This is shown by reactions 2 and 3 below. When the superoxide anion is oxidized by the catalyst, it produces neutral oxygen, O2. When the superoxide anion is reduced by the catalyst and reacts with hydrogen ions, hydrogen peroxide is produced. All SODs carry out this function to catalyze the depletion of the superoxide anions produced after the reduction of oxygen in biological systems.
|
2) 3) |
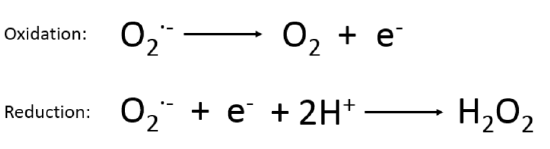 |
Antioxidant enzymes, like superoxide dismutases (SOD’s), are the primary defense against ROS. These enzymes protect the cell from damage by neutralizing ROS.3 Three primary types of SODs are found in living things and are named for the metal ion(s) found at their active sites. These enzymes also differ by the organisms and cellular locations in which they are found. The Cu/Zn superoxide dismutase (Cu/ZnSOD), is found primarily in the cytosol, nucleus, peroxisome, and intermembrane of mitochondria in eukaryotes, but can also be found in pathogenic bacteria.3 Mn superoxide dismutase (MnSOD), is found in both prokaryotes and the mitochondria of eukaryotes.1 Fe superoxide dismutase (FeSOD), is found primarily in bacteria.1 Based on the dismutation process by SODs, each form (Cu/ZnSOD, FeSOD, etc) requires a redox transition metal in order to function properly.3 MnSOD and FeSOD have similar structures, including the same ligands and geometry around the redox transition metal. This page will focus on MnSOD.
MnSOD is a Redox Catalyst
The Mn superoxide dismutase (MnSOD) is the only SOD enzyme located in the mitochondrial matrix. Mitochondria are the location of many biological processes reducing O2, producing an influx of O2˙radicals.1,3 The cofactor of the MnSOD protein is manganese (Mn).1 The Mn changes between its 3+ and 2+ oxidation states within the dismutase mechanism, consisting of two half-reactions.1 Electron exchange in metal complexes can happen by either an inner-sphere or outer-sphere mechanism. In inner-sphere electron transfer, the metal cofactor is oxidized or reduced by a ligand bound directly to the metal. In an outer-sphere mechanism, the metal is oxidized or reduced by a group not directly coordinated to the metal cofactor. The catalysis of superoxide anions by MnSOD occurs by an inner-sphere mechanism, as shown in Figure 3. The superoxide anion, depicted in red, directly binds to the available coordination site of the manganese metal cofactor.
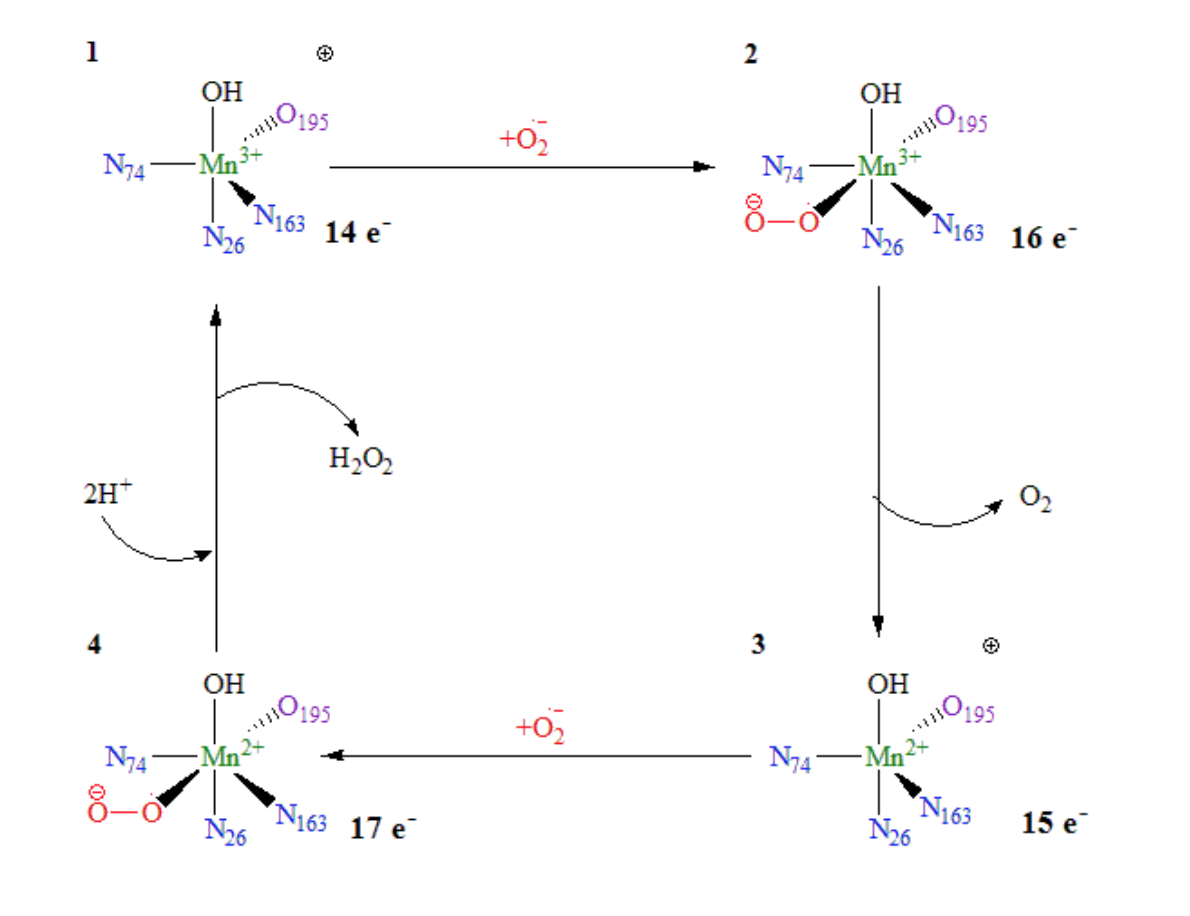
Figure 1: Proposed mechanism of dismutase of superoxides by MnSOD. Adapted by Azadmanesh et al.11 Blue nitrogens are part of the three histidine ligands. The purple oxygen indicates aspartate ligand. The superoxide anion (red) binds directly to Mn metal centers (green) by inner-sphere redox. Electron count is depicted for each intermediate. Intermediate 1 to 2: Superoxide binds to the Mn3+SOD complex. The electron from the oxidize superoxide is transferred to the reduced Mn3+, producing Mn2+SOD and O2. Intermediate 3 to 4: Electron from oxidized Mn2+SOD is transferred to the reduced superoxide, and when reacted with two hydrogen ions, produces hydrogen peroxide and Mn3+SOD.
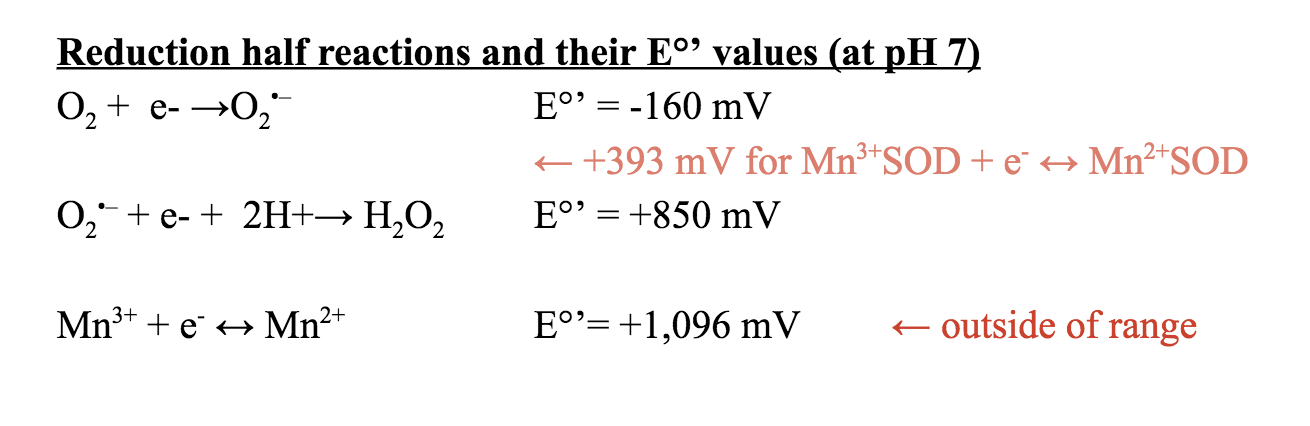
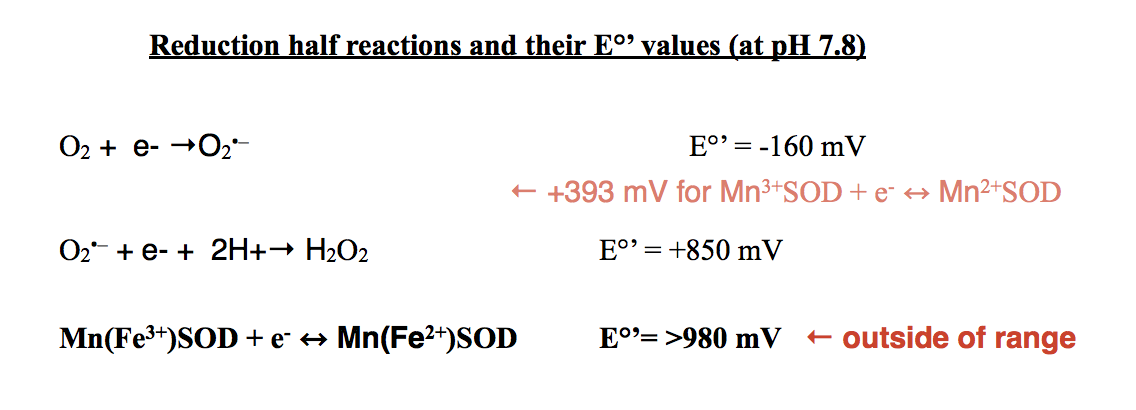
The tendency for molecules to gain electrons is measured as the redox potential, which is related to its thermodynamic stability, reaction rates, and Gibbs free energy. The redox potential, Eo, is +1,510 mV for Mn3+ + e- ↔ Mn2+.7 The E°’ value for this redox at a pH of 7 is +1,096 mV. The average redox potential (Eo’) for the MnSOD protein has been found to be 393 ± 29 mV at pH 7.7 This values lies between the E°’ values of the oxidation of superoxide to oxygen and the reduction of superoxide to hydrogen peroxide as depicted above. Due to this characteristic, the MnSOD protein is thermodynamically favorable for the catalysis of superoxide to O2 and H2O2.7
The available coordination site as well as the metal valence electrons allow for superoxide anions to bind the metal ions of the MnSOD protein. In order for a metal ion complex to be thermodynamically stable, the complexes need 18 electrons in their valence shell. As shown in the mechanism (Figure 3), when a superoxide anion is not bound to Mn3+, it has 14 total electrons in its valence shell. An additional ligand bound, which would donate x number of electrons, to the metal would make the complex more thermodynamically stable. This is the same for Mn2+. When not bound to a superoxide anion, the metal complex has 15 electrons in its valence shell. A superoxide anion bound to both metal ions results in Mn3+ ion with 16 electrons in its valence shell and an Mn2+ ion with 17 electrons. Although the electron count is not 18, it is much more thermodynamically stable than the two unbound states.
A superoxide anion is not the only compound that can bind to the MnSOD active sites as a result of the available coordination sites and electron counts. Based on experimentation when an inhibitor ligand binds to the Mn metal ion, it prevents protein functionality. Such inhibitors include an azide, fluoride, or an additional hydroxide found in solution. When the inhibitor ligand is bound, the electron count of the complex is either 16 or 17, which prevents a superoxide from binding to the active site, inhibiting the dismutase of the anion by MnSOD.
Manganese Superoxide Dismutase Structure
MnSOD is initially encoded by genomic DNA, which is upregulated by oxidative stress.5 The eukaryotic MnSOD protein is a tetramer, with four, 223 amino acid monomers (Figure 2).5 These monomers each contain N- and C-terminal domains, which pack tightly together, resembling a dimeric structure shown in Figure 5.1 Each monomer contains an active site bound to a Mn3+ ion. The monomers inherently form interfaces, increasing the overall stability of the protein’s active sites.1
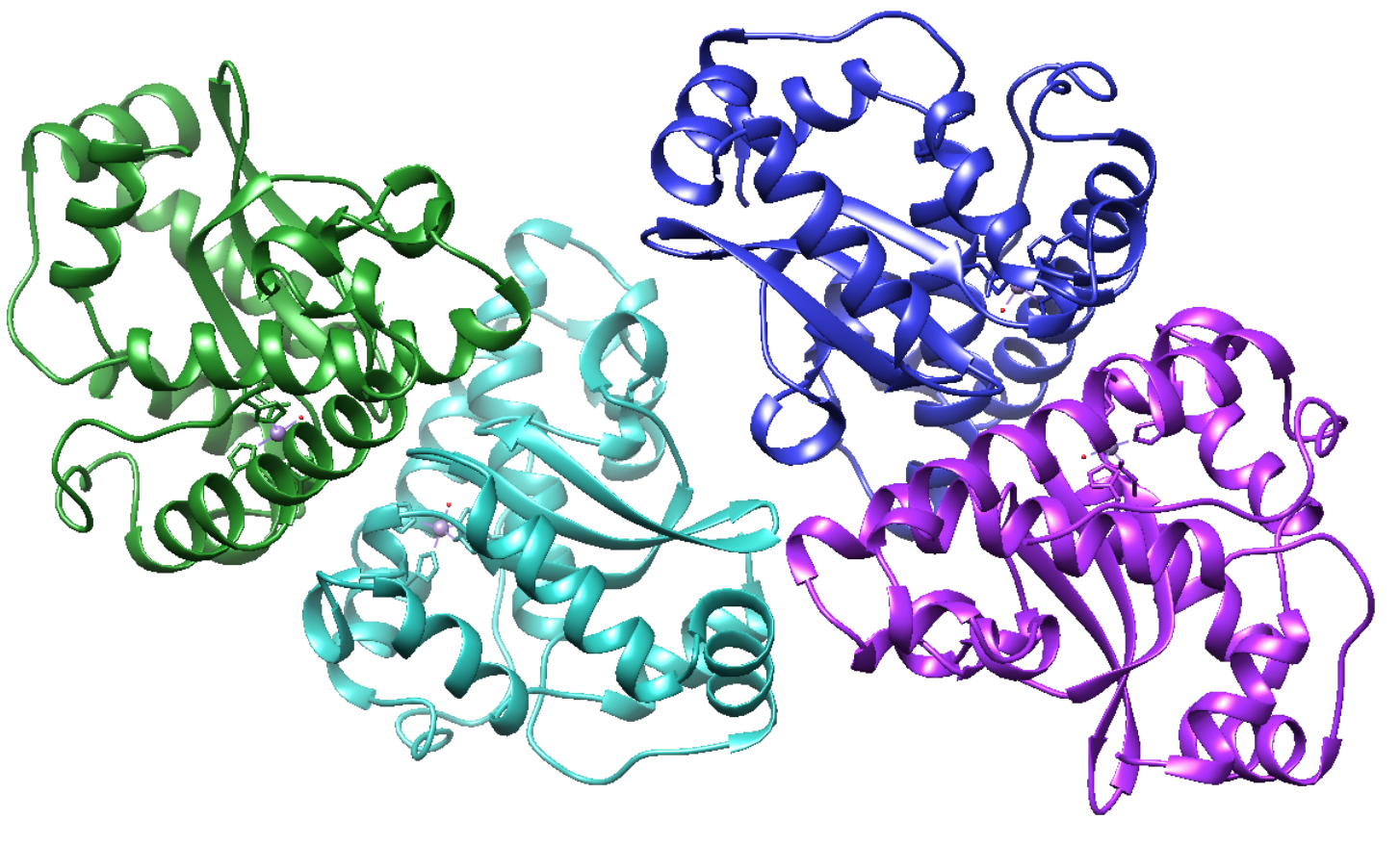
Figure 2: Overall quaternary structure of MnSOD. Adapted using Chimera PBD 1VEW. Each monomer is depicted in a different color.
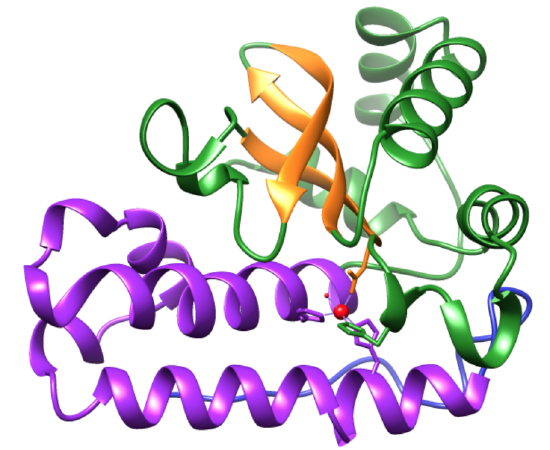
Figure 3: Each monomer contains an active site between its N- and C-termini. Two His residues from the N-terminal domain (purple) and an Asp and His residue from the C-terminal domain (green and orange) compose the metal-center active site of MnSOD.
MnSOD Active Site Selectivity
MnSOD obtains manganese ions as Mn3+ from the intracellular region of the mitochondrial matrix.4 Hard-Soft Acid-Base Theory (HSAB), ionic size and coordination state can describe why both ions can bind to MnSOD in solution. HSAB categorizes metals (acids) and ligands (bases) based on their character. The character is partly dependent on the atom’s charge density as well as on the interaction taking place. Based on HSAB, hard acid metals tend to bind to hard base ligands, with more electrostatic character, and soft acid metals tend to bind to soft base ligands, resulting in bonds with more covalent character. Borderline metal acids and ligand bases demonstrate intermediate character. Both Fe3+ and Mn3+ are hard acids. As depicted in Figure 6, aspartate and hydroxide are hard bases. This means that they tend to interact with hard acids with more electrostatic character. Histidine on the other hand, is a borderline base. However, the Asp and hydroxide ligands are hard bases, the protein active sites tend to bind with hard acids like Fe3+ and Mn3+. Thus the metal-ligand interactions in the active site follow HSAB Theory thermodynamics.
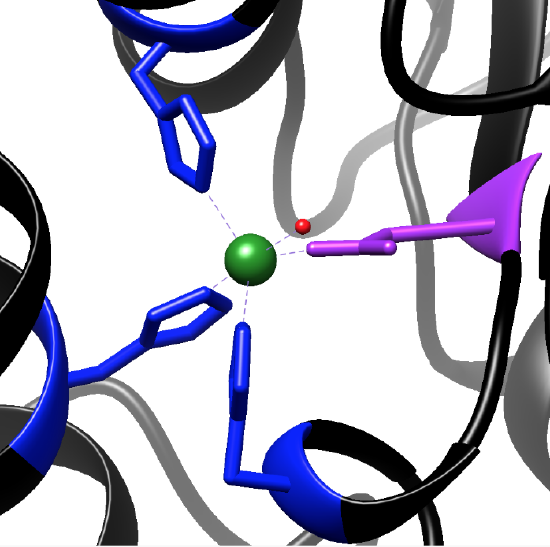
Figure 4: MnSOD active site with Mn (green) bound to three His (blue), an Asp (purple), and an oxygen atom (red) from hydroxide. Figure adapted from Chimera using PDB 1VEW.
The MnSOD active sites bind only to the 3+ oxidation state of each metal ion.4 The ionic size of Fe3+ and Mn3+ are identical based on Table 1, indicating that it may be highly probable that the active site of MnSOD is size-specific.
Table 1: Ionic Radii and Spin States of metal ions that bind to MnSOD11
It has been determined that the MnSOD active site when bound to Fe3+ is identical to the active site of bound Mn3+.8 Thus, they both demonstrate a five-coordinate geometry. However, experimentation has shown that in E. coli, MnSOD is active when Mn3+ is bound, but not when Fe3+ is bound.1,4 When bound to MnSOD, Mn3+ has a specific activity of about 4,700 units/mg.4 In comparison, when Fe3+ was bound to the MnSOD protein, the specific activity was about 140 units/mg.4 MnSOD, with Fe3+ as its metal cofactor, is inactive because of its redox potential. Recall that the redox potential of MnSOD with Mn3+/2+ lies between the E°’ values of the oxidation of superoxide to oxygen and the reduction of superoxide to hydrogen peroxide. This character has been found to be vital for MnSOD function. Mn(Fe3+/2+)SOD redox potential is too high for catalytic turnover as depicted below.8 Therefore, MnSOD binds to both Mn3+ and Fe3+, but is only active with Mn3+/2+.
Importance of Geometry
The geometry of the functional active site of MnSOD at room temperature is a five-coordinate, trigonal bipyramidal geometry (Figure 5).1 This leaves an open coordination site on the metal for a superoxide anion to bind. The bound ligands include three histidines (His26, His74, His163), an aspartate (Asp195), and a single hydroxide molecule.1 The histidine and aspartate residues are bound to the Mn ion through the protein backbone. Thus, each active site is multidentate, resulting in high stability when bound with the Mn metal ions.
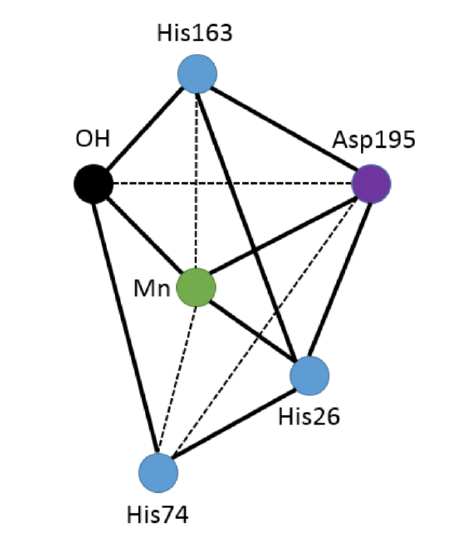
Figure 5: Adapted from Borgstahl et. al.3
Ligand Field Theory, LFT, depends on the geometry of the metal complex. LFT is the application of molecular orbital (MO) theory to metal coordination complexes. For metal coordination complexes, LFT helps explain the bonding, nonbonding, and antibonding orbitals that arise from interaction of the ligand orbitals and the d-orbitals of the metal ion. The splitting energy (Δ) of the complex is the energy difference between the bonding and antibonding orbitals of the molecule. When the electrostatic repulsion energy of pairing electrons is greater than the Δ of the orbitals, the complex is high spin. High spin complexes tend to fill higher energy orbitals with valence electrons, specifically d orbital electrons, before the electrons pair in lower energy orbitals. This is because it requires less energy to overcome the Δ of the complex than the electrostatic repulsion energy of the electrons. When the electrostatic repulsion energy of the electrons is less than the Δ of the complex, the electrons pair in lower energy orbitals before filling higher energy orbitals. The energies and the Δ of the orbitals depends on the geometry of the molecule as well as the identities of both the ligands and the metal.
The metal ion of the complex has a large effect of the splitting energy of the system. Metal complexes can demonstrate either high or low spin states. 3d transition metals can demonstrate either high or low spin. Metals with a larger charge tend to result in a higher Δ. Thus, 3+ metals tend to be high spin. For 4d and 5d transition metals, Δ tends to be larger than the Δ for 3d metals. Thus, these complexes are almost always low spin. Mn3+ is high spin based on its charge and Mn2+ can be either high or low spin. To determine the spin state if the complexes, we can look to the ligands on the active site for further information.
The ligands of a complex can affect the Δ based on the nature of their bonding interactions. σ donor ligands are ligands that have one pair of electrons on the donor atom. Under this category of ligands, the stronger the base, the larger the Δ. Like explained before, if Δ is large, the complex tends to be low spin. Low spin ligands are also categorized as strong field. In contrast, weak field ligands are also high spin. σ donor and π donor ligands have two or more pairs of electron on the donor atom and demonstrate a weak field. This means that the π donating character of the ligand decreases Δ. As a result, σ donor and π donor ligands tend to demonstrate high spin states. σ donor and π acceptor ligands contain one pair of electrons, a π* orbital on the donor atom, and demonstrate a strong field. These ligands tend to increase Δ and encourage low spin as a result.
The Asp carboxylate and hydroxide ligands of the MnSOD active site are σ and π donating. This indicates it is a weak field ligand, promoting high spin. Histidine imidazole is considered σ donating and π accepting, which can result in a strong field and low spin with a large Δ . However, the combination of the high spin character of Mn3+, Asp, and hydroxide the overall metal complex is high spin.8
Active Site Stability
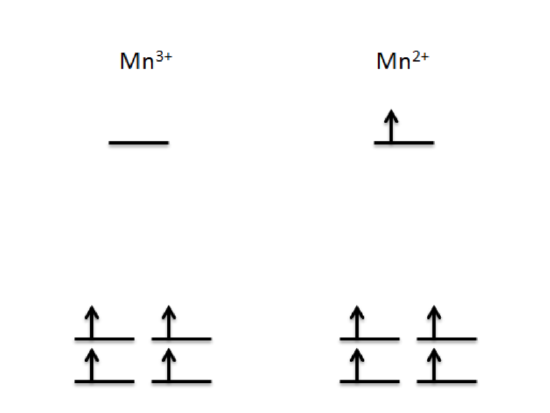
Figure 6: Trigonal bipyramidal splitting diagrams of both Mn3+ and Mn2+, the two utilized manganese oxidation states found in MnSOD active sites.
Since both Mn3+ and Mn2+ demonstrate high spin states, the splitting diagrams of each oxidation state demonstrate the d-orbital electrons fill each orbital before doubling up. Figure 7 depicts the trigonal bipyramidal splitting energy diagrams respectively. This depiction shows that Mn3+, has 4 d electrons occupying lower energy d orbitals and Mn2+ with an additional electron in a higher energy d orbital. Based on Ligand Field Stabilization Energy (LFSE), the complex with fewer electrons in high energy orbitals have higher thermodynamic stability. In addition, due to the larger charge of Mn3+, the greater amount of protons in the metal complex than electrons result in a strong pull on the ligands. Thus, the Mn3+ complex has a greater Zeff, so it is more stable than the Mn2+ complex.
The more stable the metal complex, the less labile it is. Lability, or reactivity, is defined as the rate in which ligands are replaced in coordination complexes. This rate of ligand exchange depends mostly on electrostatics and electron filling in d-orbitals. When a metal center is labile, the rate in which ligands are replaced in coordination complexes is high. The ligand replacement rate in inert complexes is very slow if any exchange occurs at all. Since the Mn3+ complex is more stable than the Mn2+ complex, it is more inert. However, the lability of both complexes play a less important role in the binding of superoxide anions compared to the metal ion coordination and valence electron stability.
Electronic Absorptions within MnSOD
Both Mn3+ and Mn2+ have available d orbitals for d-d or charge-transfer transitions. The human MnSOD protein itself demonstrates absorbance on the visible spectrum at 480 nm.6 Thus, the MnSOD protein emits a red-orange color. The absorbance of the MnSOD protein heavily depends on the pH of the system. As the pH of the system increased to about 9.4, the system was “bleached”, or the absorbance was less intense.6 Thus the ϵ value decreased, with a maximum ϵ of 525 M-1 cm-1 at a pH of about 7.8.6 The pH change may have affected the protein active site structure, resulting in a less intense absorbance.
With an ϵ of 525 M-1 cm-1, available d-orbitals, π-donating ligands, and heavy dependence on pH, it can be hypothesized that the MnSOD protein demonstrates ligand-to-metal charge-transfer (LMCT) transitions. The absorbed energy excites electrons within the ligands to empty d orbitals. The available d orbitals for these excited electrons can be found in the splitting orbitals diagrams of both Mn3+ and Mn2+ in Figure 7.
Summary
Every aspect of the MnSOD protein is vital for biological function. The protein, along with other SOD protein, help combat against ROS, preventing oxidative stress and diseases such as Alzheimer’s, apoptosis in the brain, and diabetic cardiopathy.3
- Borgstahl, G. E. O.; Parge, H. E.; Hickey, M. J.; Beyer Jr., W. F.; Hallewell, R. A.; Tainer, J. A. The Structure of Human Mitochondrial Manganese. e Superoxide Dismutase Reveals a Novel Tetrameric Interface of Two 4-Helix Bundles. Cell, 1992; Vol 71, pp 107-118
- Borgstahl, G. E. O.; Pokross, M.; Chehab, R.; Sekher, A.; Snell, E. H. Cryo-Trapping the Six-Coordinate, Distorted-Octahedral Active Site of Manganese Superoxide dismutase Edited by R. Huber. Journal of Molecular Biology 2000, 296 (4), 951–959.
- Bresciani, G.; da Cruz, I. B. M.; González-Gallego, J. Chapter Four - Manganese Superoxide Dismutase and Oxidative Stress Modulation. In Advances in Clinical Chemistry; Makowski, G. S., Ed.; Elsevier, 2015; Vol. 68, pp 87–130.
- Beyer, W. F; Fridovich, I. In Vivo Competition between Iron and Manganese of Occupancy of the Active Site Region of the Manganese-Superoxide Dismutase of Escherichia coli. The Journal of Biological Chemistry 1991, Vol. 266, 303-308
- Candas, D.; Li, J. J. MnSOD in Oxidative Stress Response-Potential Regulation via Mitochondrial Protein Influx. Antioxid Redox Signal 2014, 20 (10), 1599–1617.
- Hsu, J.-L.; Hsieh, Y.; Tu, C.; O’Connor, D.; Nick, H. S.; Silverman, D. N. Catalytic Properties of Human Manganese Superoxide Dismutase. J. Biol. Chem. 1996, 271 (30), 17687–17691.
- Leveque, V. J.-P.; Vance, C. K.; Nick, H. S.; Silverman, D. N.; Redox Properties of Human Manganese Superoxide Dismutase and Active-Site Mutants. Biochemistry 2001, 40, 10586-10591
- Jackson, T. A.; Gutman, C. T.; Maliekal, J.; Miller, A.-F.; Brunold, T. C. Geometric and Electronic Structures of Manganese-Substituted Iron Superoxide Dismutase. Inorg Chem 2013, 52 (6), 3356–3367.
- Srnec, M.; Aquilante, F.; Ryde, U.; Rulisek, L. Reaction Mechanism of Manganese Superoxide Dismutase Studied by Combined Quantum and Molecular Mechanical Calculations and Multiconfigurational Methods. J. Phys. Chem. B 2009, 113, 6074-6086
- Whittaker, M. M.; Mizuno, K.; Bächinger, H. P.; Whittaker, J. W. Kinetic Analysis of the Metal Binding Mechanism of Escherichia Coli Manganese Superoxide Dismutase. Biophys J 2006, 90 (2), 598–607.
- Azadmanesh, J.; Borgstahl, G. E. O.; A Review of the Catalytic Mechanism of human Manganese Superoxide Dismutase, Antioxidants 2018
- Atomistic Simulation Group in the Materials Department of Imperial College; Database of Ionic Radii; http://abulafia.mt.ic.ac.uk/shannon/ptable.php
Contributed By:
This work was originally written by Morgan Matthews, Spring 2018: Morgan is currently (as of 2018) a senior chemistry major at Saint Mary's College in Notre Dame, IN, and will graduate from the University of Notre Dame with her dual degree in Chemical Engineering next spring.
This work was originally edited by Dr. Kathryn Haas (Assistant Professor), Madison Sendzik (Teaching and Research Assistant), and Dr. Dorothy Feigl (Professor) at Saint Mary's College.

