Keep Cu Safe: Intracellular Copper Chaperones
- Page ID
- 68250
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Copper Chaperone Atx1
Copper chaperones are intracellular proteins that protect Cu along the pathway from Cu entry into the cell, through the cytoplasm, to its intracellular targets. This page focuses on the yeast Cu chaperone Atx1. Intracellular Cu chaperones, like Atx1, are well-conserved throughout eukaryotic species. The analogous protein in mammalian cells is Atox1.
This is a video introduction to the intracellular yeast Cu chaperone protein Atx1 (or Atox1 in mammals).
A copper chaperone is protein which binds intracellular copper and delivers it safely to essential intracellular locations. The primary function of a copper chaperone protein is to prevent unwanted, and dangerous, oxidizing reactions of copper as it travels through the cytoplasm. Unwanted redox cycling of copper causes a chain of aberrant chemical reactions that occur in the cell when unregulated Cu(I) interacts with oxidative species and then produces Cu(II) and reactive oxygen species, or ROS.1 The reactive oxygen species, such as HO⦁-, are hazardous to the health of the cell because the potential impact of oxidative stress when they interact with proteins and surfaces within the cell, overall leading to the aging and potential death of the cell. Figure 1 below provides a scheme for redox cycling in the cell. This reaction is continually harmful because the Cu(II) may be reduced to Cu(I) and the cycle continues to cause cell damage as long as the metal remains free. If copper chaperones do not properly transport and protect intracellular copper, the reactive products of redox cycling will irreversibly damage the cell. In order to prevent redox cycling, the copper chaperone stabilizes one oxidative state of copper; the Cu(I) oxidation state is selected because the cell is a reducing environment and prefers this case already.1
Redox cycling occurs when reactive products of metabolism meet free copper to produce oxidative species that may cause damages to the cell. (Original artwork by Adrienne Bruggeman)

This page focuses on the yeast copper chaperone, Antioxidant1, or Atx1 in shorthand. Copper chaperone Atox1 is the homologous chaperone present in mammalian (human) cells.2 Yeast (Saccharomyces cerevisiae) is a model system that is simpler to study than mammalian cells. Most of what we know about the copper transport pathways of mammalian cells is derived from yeast model studies. Atx1 in yeast is assumed to be identical in its function to the human protein, Atox1.2 These proteins are part of an elaborate system dedicated to safe copper transport through the cellular cytoplasm. This system protects the cell by relaying copper from the plasma membrane protein Copper Transporter 1 (Ctr1), to Atx1, to the ATP-dependent Cross-Complements Ca2+ protein, abbreviated Ccc2, which takes Cu into the golgi.3 In short, the Atx1 is a member of a team of proteins dedicated to copper transport, protection, and usage through the cell. In mammalian cells, the copper chaperone Atox1 accepts copper from hCtr1 and delivers to proteins ATP7A and ATP7B. Atx1 and all copper chaperones are integral features of copper transport within the cell, most importantly stabilizing the Cu(I) oxidation state in order to prevent the harm caused by redox cycling.
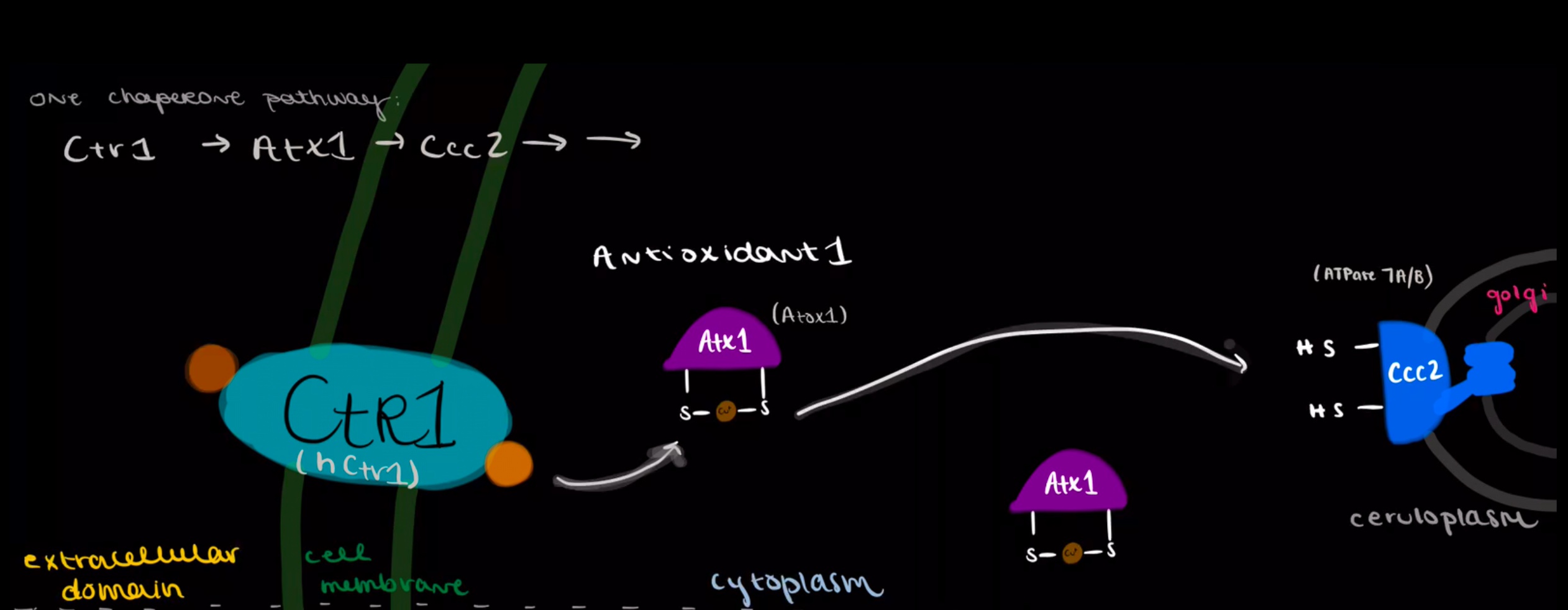
The figure below illustrates one of the yeast intracellular Cu pathways which is mediated by the Cu Chaperone Atx1 (or Atox1 in mammals). Atx1 shuttles Cu from its cellular entry point at Ctr1 to the golgi, where it can be incorporated into enzymes destined for outside the cell. Ctr1 is the plasma membrane Cu transporter that is responsible for transporting Cu from the extracellular environment to the cytoplasm (hCtr1 in human). Ccc2 is the ATP-dependant protein that recieves Cu from Atx1 and transports Cu into the golgi. ATPase7A and ATPase7B are the human analogs to yeast Ccc2. (Original artwork by Adrienne Bruggeman)
Hard soft acid base (HSAB) theory can be applied to explain how the chaperone’s donor atoms help to stabilize Cu(I) over Cu(II) to prevent redox cycling. The theory of HSAB categorizes acids and bases according to ionic radius, charge/ion balance, and charge of ions to predict the binding capacity between an acid and a base.4 The HSAB theory follows the convention that soft acids and bases prefer to interact while hard acids and bases prefer to interact compared to a hard-soft mixed interaction. According to this theory, the thiolate (S-) ligand of a deprotonated cysteine is a soft base due to its large ionic radius and small charge. Similarly due to radius and charge, Cu(I) is characterized as a soft acid, while Cu(II) is characterized as a borderline acid. Thus, the soft binding site of Atx1 prefers to bind with the soft Cu(I) rather than the borderline Cu(II), and in the presence of both, the sulfur will prefer the Cu(I) and not the Cu(II). This strong preference for Cu(I) over Cu(II) increases the stability of Cu(I) in the cell, and helps prevent it from being oxidized to Cu(II). This stabilization of Cu(I) discourages undesirable redox cycling.
Ligand Field Theory (LFT) is another principle of coordination chemistry that can be applied to explain how Atx1 stabilizes the Cu(I) oxidation state. The d10 electron filling of Cu(I) results in a ligand field stabilization energy (LFSE) of zero.5 The value of zero comes from the balance of a full set of electrons in higher energy orbitals and in lower energy orbitals. A zero LFSE indicates that there is no stabilization energy for any particular geometry, and thus gives the Cu(I) freedom to bind in any geometry.2 When bound to Atx1, Cu(I) takes a linear geometry between two cysteine residues. Cu(II) is a d9 metal with a LFSE of -0.6Δ. This negative stabilization energy indicates that the Cu(II) will have geometric preference for octahedral-type geometry, and thus would disfavor 2-coordinate binding geometries.5 Thus the geometry of the Atx1 binding site is disfavorable to Cu(II), further stabilizing the preference of Cu(I) in the cell.
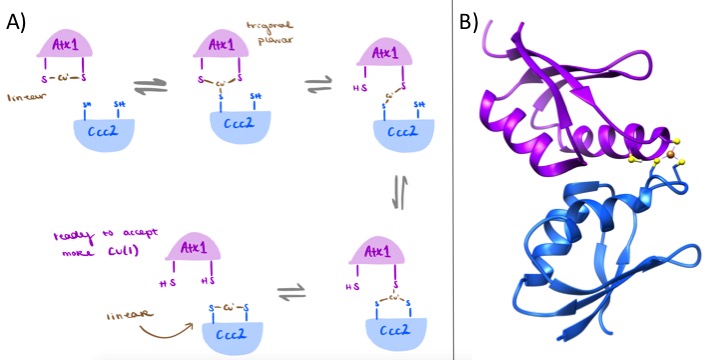
The zero LFSE of Cu(I) also assists in the transfer mechanism from the copper chaperone to another protein, such as Ccc2. During the docking and transfer process to Ccc2, copper bound to Atx1 forms an additional bond with an adjacent cysteine residue of the docked Ccc2 protein before breaking any initial bonds with Atx1.3 Part A of the figure below provides an illustration of the transfer mechanism from Atx1 to Ccc2. Note that the geometry transitions from linear to trigonal planar before breaking the bonds with Atx1 and returning to a linear geometry at the Ccc2 site.3 The two-coordinate linear and three-coordinate trigonal planar complexes are both favorable to the Cu(I) over Cu(II) due to the LFSE preference. This mechanism again promotes stability of Cu(I) and disfavors Cu(II).
A) The cartoon diagram of the Atx1 transfer of copper to Ccc2. The complex begins as 2-coordinate, becomes 3-coordinate twice during the transfer and ends as 2-coordinate again. (Original artwork by Adrienne Bruggeman) B) A ribbon diagram of the three-coordinate complex of Atx1 (purple) to Ccc2 (blue).
C) Below is an animation of the transfer mechanism illustrated above.
The Cu chaperone transfer mechanism also makes sense in terms of electron counting (think 18-electron rule). As previously mentioned, Cu(I) is a full shell d10 metal and is bound to two sulfurs of the Atx1 chaperone. Each of the sulfur ligands will donate two electrons to the copper. Thus the total electron count of the metal binding site is 14. A metal’s capacity for electrons is 18, and since the copper does not yet have 18, it is able to accept electrons until it reaches this capacity. This ability to accept electrons is relevant to the copper chaperone system because at its binding step, which docks the copper to Ccc2, the copper accepts an additional sulfur bond before breaking either of the preexisting cysteine bonds with Atx1. Cu(I)’s ability to accept an additional ligand from Ccc2 while it is still coordinated to Atx1 means that that the metal will not be released as a free ion into the cell.3 Thus Cu is always “protected” by Atx1 and/or Ccc2 coordination, preventing redox cycling.
The LFSE and geometry-fluidity of Cu(I) not only impacts the transfer mechanism towards greater stability of Cu(I) over Cu(II), but also reduces the thermodynamic energy cost during the transfer of Cu between Atx1 and Ccc2. Through the geometry-fluidity of Cu(I), the metal will bind to the chaperone reversibly and easily change its geometry when necessary. Thus at the end of the mechanism, an unbound Atx1 is produced.6 If Cu(I) tended to bind with a specific preferred geometry, it would be more difficult to change out of that geometry. Thus the zero LFSE allows the Cu(I) to change geometry with low thermodynamic cost.
Finally, the lability of the Cu(I) reduces the energy cost of the transport from Atx1 to Ccc2. When a metal ion is labile, this means that it reacts at a high kinetic rate and may readily replace ligands in coordination complexes.7 Copper, both as Cu(II) and as Cu(I) is especially labile. This lability comes as a result of the presence of d-electrons in the complex's high energy antibonding orbitals. When these electrons are present in the high energy orbitals, they contribute to high lability (the complex exchanges ligands at a high rate). When the Cu(I) enters the cell through Ctr1, the copper chaperone complex binds and transports the copper through the cell to the metal’s next binding site. The copper’s high lability is necessary so that the metal is able to change ligands readily, thus encouraging the ongoing binding and transport of the metal between chaperones and target proteins.7
The protective function of the copper chaperone is realized by the application of many chemical principles towards the prevention of redox cycling in the cell. In order to best prevent redox cycling, Atx1 stabilizes one oxidation state of copper. Because the cell is a reducing environment, it prefers Cu(I) over Cu(II), and because Ctr1 delivers copper in this form into the cell, the copper chaperone Atx1 is best suited to stabilize Cu(I). According to HSAB Theory and Ligand Field Theory, the binding site of Atx1, composed of two linear cysteines, stabilizes Cu(I) over Cu(II). Similarly, the mechanism of copper transfer from Atx1 to Ccc2 is used to further stabilize Cu(I) over Cu(II) in the cell and to prevent the exposure of free copper to the cell. Through coordination geometries only accessible to Cu(I) and the high lability of copper, the copper chaperone is able to transfer copper stably and with low energy costs. The ability for Atx1 and any copper chaperone to bind stably with copper not only successfully transports biologically needed metal ions, but also proactively reduces the damaging effects of free copper, redox cycling, and oxidative stress to the cell.
Sources
- Rodriguez-Montelongo, L.; de la Cruz-Rodriguez, L. C.; Farías, R. N.; Massa, E. M. Membrane-Associated Redox Cycling of Copper Mediates Hydroperoxide Toxicity in Escherichia Coli. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1993, 1144 (1), 77–84.
- Rosenzweig, A. C.; O’Halloran, T. V. Structure and Chemistry of the Copper Chaperone Proteins. Curr Opin Chem Biol 2000, 4 (2), 140–147.
- Finney, L. A.; O’Halloran, T. V. Transition Metal Speciation in the Cell: Insights from the Chemistry of Metal Ion Receptors. Science 2003, 300 (5621), 931–936.
- Gray, H.B.; Stiefel, E. I.; Valentine, J.S.; Bertini, I. Biological Inorganic Chemistry: Structure and Reactivity. 2007. 331-337.
- Schaller, C. P. Ligand Field Theory http://chem.libretexts.org/LibreTexts/Saint_Mary’s_College%2C_ Notre_Dame%2C_IN/CHEM_342%3A_Bio-inorganic_Chemistry/Chapters/Chapter_3%3A_Introduction_to_Advanced_Bonding_Theories/3.2_Ligand_Field_Theory/B._Ligand_Field_Theory/B.1_Ligand_Field_Theory (accessed Oct 11, 2016).
- O’Halloran, T. V.; Culotta, V. C. Metallochaperones, an Intracellular Shuttle Service for Metal Ions. J. Biol. Chem. 2000, 275 (33), 25057–25060.
- Pope, C. R.; De Feo, C. J.; Unger, V. M. Cellular Distribution of Copper to Superoxide Dismutase Involves Scaffolding by Membranes. Proc. Natl. Acad. Sci. U.S.A. 2013, 110 (51), 20491–20496.
Contributed by:
This work was originally written by Adrienne Bruggeman, Fall 2016: Adrienne is currently (as of 2016) a senior chemistry major at Saint Mary's College in Notre Dame, IN.
This work was originally edited by Dr. Kathryn Haas, Assistant Professor at Saint Mary's College.

