Iron acquisition in bacteria: Siderophores
- Page ID
- 67339
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Siderophores
This is a video introduction to siderophores: the small molecules produced and secreted by bacteria to acquire precious iron from their environment.
Most all living organisms require iron for crucial processes that allow for the life and growth of cells. Animals receive this nutrient from diet, but plants and single-celled organisms must utilize different strategies.1 There are two main mechanisms by which cells acquire iron. The simpler of the two, is iron diffusion across cellular membranes.2 This mechanism is more difficult, despite the prominence of iron in the atmosphere. This is due to the aerobic nature of the environment, which causes iron to be present in the highly insoluble form, Fe(OH)3. The insolubility means there is a low Fe(III) concentration (10-18 M) of available iron; too low to provide sustenance to microorganisms, which require a minimum concentration of 10-8 M to live and grow.3, 4 To circumvent the lacking bioavailability of iron, these organisms synthesize and secrete siderophores. Siderophores are low molecular weight compounds that chelate iron and transport it into cells under low iron environments.5 Siderophores have the highest specificity and affinity for Fe(III), but are capable of forming complexes with other metallic ions, like Fe2+, Zn2+, Ga3+, and Cr3+.2
Siderophores are an incredibly diverse group of biocompounds, with hundreds of different types categorized. The most common siderophores fall into three main categories: hydroxamates, catecholates, and carboxylates.2 The functional groups of these structures that are the main component in iron chelation are shown below, in Figure 1.
Figure 12: Functional groups of siderophores and the complexes that form upon binding to Fe(III). R-groups represent the variable groups.
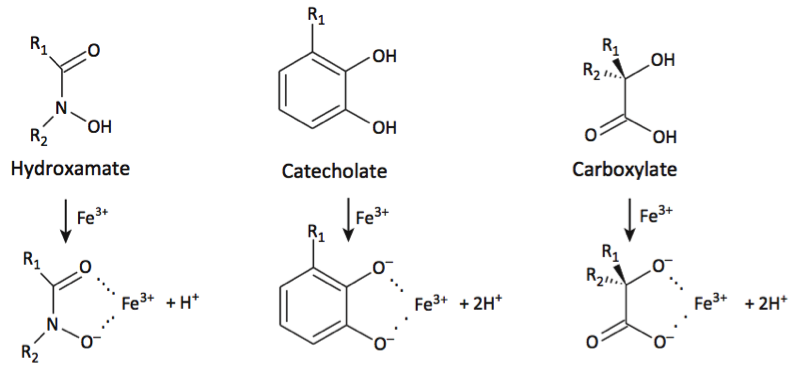
These bidentate functional groups interact with iron through two negatively-charged oxygen atoms, but this is not a steadfast rule. Some distortions can occur and other atoms like nitrogen or sulfur can be substituted as one of the interacting atoms (although this has a tendency to reduce Fe(III) affinity).6 The functional groups act as bidentate ligands, but naturally occurring siderophores can arrange multiples of these groups through the variable R-groups into a higher chelating structure. Hexadentate and tetradentate structures are the most common. Hexadentate siderophores tend to bind to Fe(III) more so than tetradentate or bidentate siderophores, due to the chelate effect. The resulting hexadentate complexes are also more stable and less labile, which makes sense, since one iron-bound ligand–instead of two or three–increases the entropy.3 Examples of hexadentate siderophores are in Figure 2.
Figure 2: Examples of the three types of siderophores in a hexadentate formation. The colored portions highlight the iron and siderophore functional groups. The light gray represents the variable groups. (A) A ferrichrome ligand, a type of hydroxamate from an Esherichia coli cell.7 (B) An enterobactin ligand, a type of catecholate from a Bacillus subtilis cell.8 (C) A staphyloferrin ligand, a type of carboxylate from a Staphylococci.9

Fe(III) is typically found in an environment with an aqueous solution, meaning the naturally-occurring complex is the octahedral Fe(H2O)63+. Siderophores surround this complex, usually in a hexadentate manner that results in octahedral geometry. Typically, the water is replaced completely by siderophore in an entropically-favorable way. In this new complex, Fe(III) is in a d5 electron configuration. As previously mentioned, siderophores donate multiple oxygen atoms to form a complex with iron. The donation of oxygen atoms means these ligands are σ and π donors, which are weak field ligands and support a high spin configuration. The combination of the complex’s geometry and the metal center’s high spin configuration means that there is no ligand field stabilization energy.
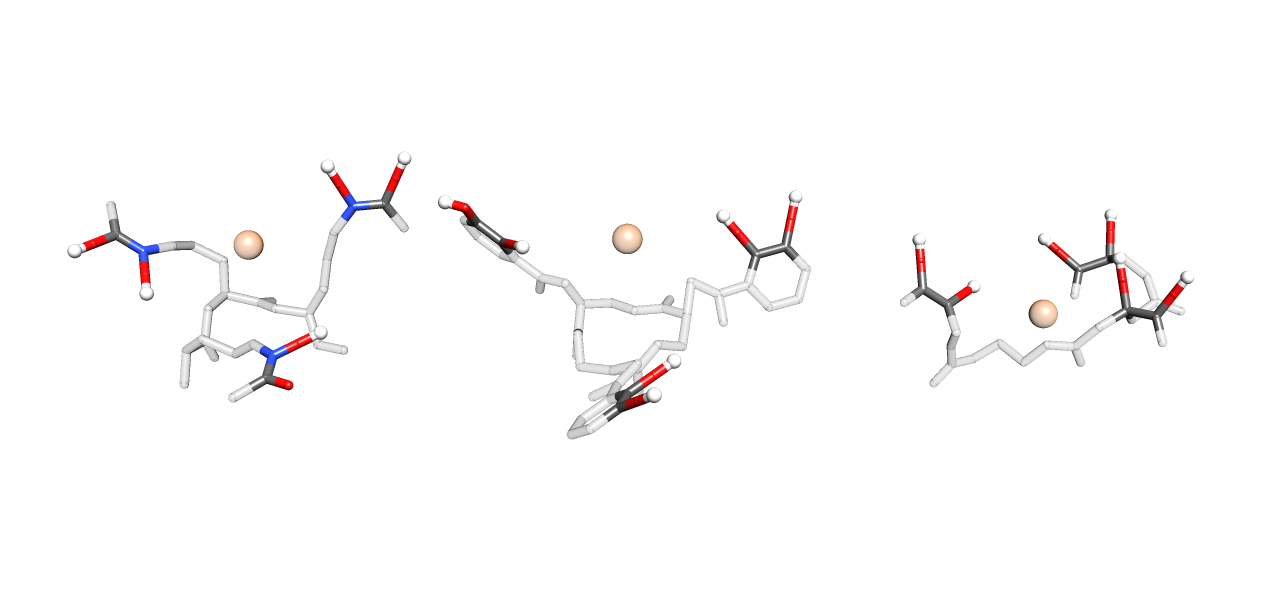
Without bound Fe(III), siderophore are not rigid compounds (Figure 3). The metal-ligand bond is what stabilizes their structure. Complex stability and the siderophores’ high affinity for iron comes from the aforementioned ligand denticity as well as the multiple hard acid and hard base interactions. Fe(III) is a hard acid and the oxygen atoms on the siderophore are all hard bases.10 The affinity for iron increases with higher negative charges on siderophores.6 More negatively-charged siderophores are harder bases, which form tighter interaction to Fe(III). As shown in Figure 1, catecholates and carboxylates usually have two negatively-charged oxygen atoms per functional group, while hydroxamates have one. This alludes to the idea that the former groups have higher iron affinity.
Fe(III)-siderophore complexation is sensitive to pH values. Free iron and protons (hydrogen atoms) compete for the unbound siderophores.11 In low pH environments, there is a high concentration of solvated hydrogen atoms, resulting in higher competition for the siderophores (Figure 3). A low pKa value relates to a strong acid that is able to donate protons. In Figure 1 above, both the catecholate and carboxylate groups donate two hydrogen atoms (protons), as opposed to the hydroxamate group, which donates one. The former two will have lower pKa, and thus a higher negative charge density, which means a higher iron affinity.6
Figure 37, 8, 9: Siderophores unbound to Fe(III). The octahedral geometry is not held without the metal. Here, hydrogens have bound to the siderophores, replacing the Fe(III) bonds.
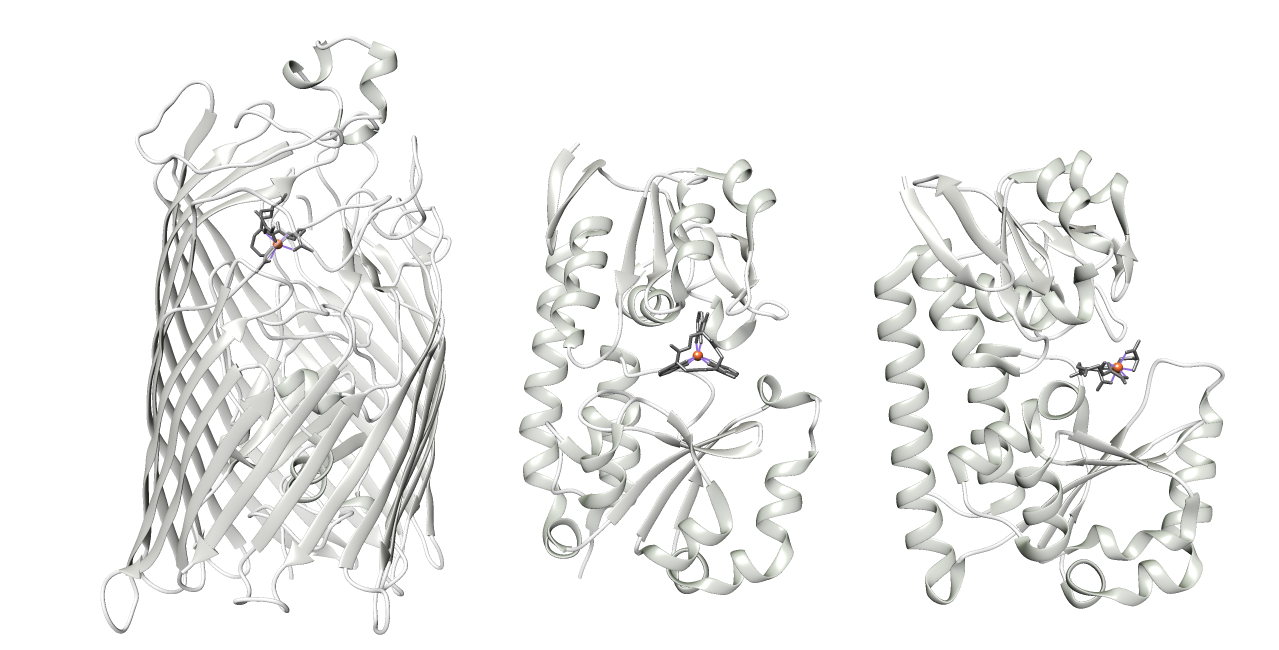
Once the Fe(III)-siderophore complex has formed, it is introduced into the cells by complex, energy-dependent transportation mechanisms.12 The introduction usually begins when the complex binds to a receptor protein on the cellular outer membrane, which have high affinity for the complexes. The binding changes the protein conformationally as the Fe(III)-siderophore complex is shuttled through and released into the periplasm. Once there, it binds to another high affinity proton and then taken directly or passed through a chain of transport proteins until it reaches its final destination.13 Examples of some membrane-bound transport proteins are in Figure 4.
Figure 4: Examples of the three types of siderophores in a hexadentate formation. The dark gray and colored portions highlight the Fe(III)-siderophore complexes. The light gray represents the membrane proteins. (A) An FhuA protein bound to a ferrichrome ligand.7, 12 (B) An FeuA protein bound to an enterobactin ligand.8, 14 (C) A SirA protein bound to a staphylococci ligand.9, 15
The function of a siderophore is to bring insoluble iron from the environment into the cell. All siderophore perform this task uniformly. However, there are multiple mechanisms for iron release into the cell. There are two common mechanisms utilized by the majority siderophores. The first involves a reduction of Fe(III) to Fe(II), which results in a lowered iron affinity and looser bond to the siderophore. This is energetically favorable as it does not degrade the siderophore in any way, so it can be reused in iron acquisition.16 Less frequently, the backbone of a siderophore undergoes a hydrolysis reaction. These have a tendency to yield a siderophore product that is useless for re-secretion into the environment.16, 17
Reduction to Fe(II) produces a weaker bond to the siderophore and, overall, a less stable complex. Stability constants for the reduced iron form are on average 20 orders of magnitude lower. Contributing to this, is the fact that Fe(II) is a softer acid, which has weaker interactions with the strongly basic oxygen-donor groups. What’s more, it’s not as effective at outcompeting solvated hydrogens to bind these groups. This weaker metal-ligand interaction means Fe(II) is more labile and likely to dissociate.3
The stability of the Fe(III)-siderophore complexes make it difficult for reduction to occur, but various facilitators help the reaction along. There are certain chelators and reductive enzymes that directly catalyze the reduction reaction, but the surrounding environment plays multiple roles as well.6 Hydrophobic regions and low pH mediums stimulate reduction. In acidic conditions, H+ ions can partially dechelate the iron-siderophore complex. This reduced ligand denticity results in a less stable complex, which makes reduction more likely.3
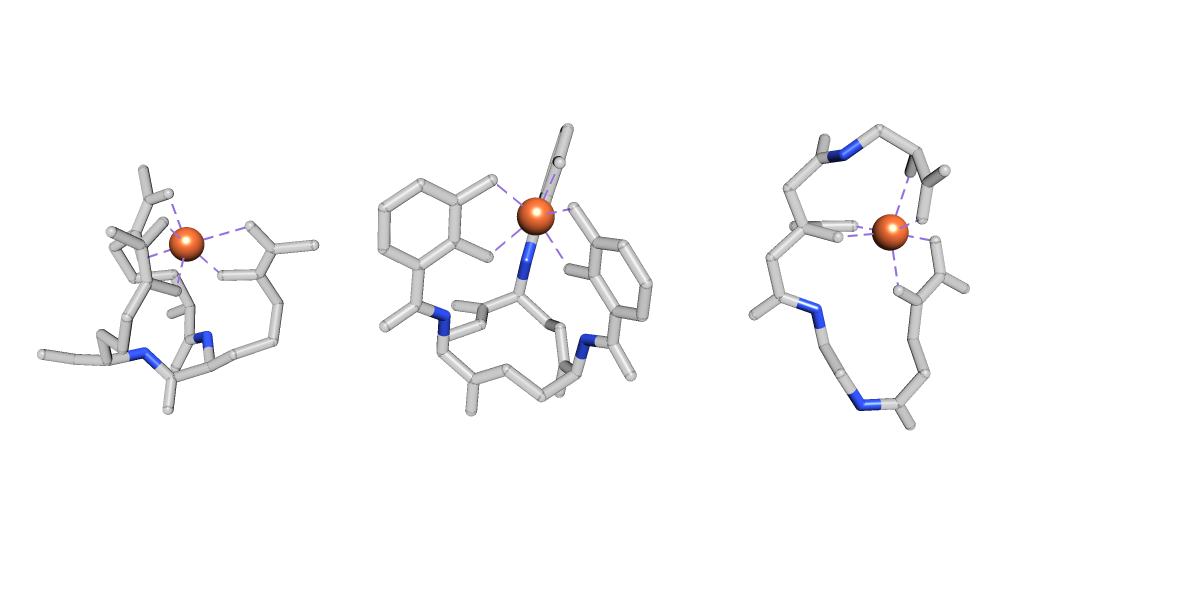
Reduced denticity is the main theory behind the second release mechanism. Here chelation is decreased through hydrolysis of the siderophore backbone instead of through protonation of the donor groups. This reaction effectively breaks the siderophore, but significantly decreases iron affinity. The disposal of a siderophore is not energetically favorable, which is one reason this mechanism is not as popular of a strategy. What’s more, siderophore hydrolysis requires a siderophore with hydrolyzable groups embedded into its backbone (Figure 5), which leaves a limited selection of siderophores.3
Figure 57, 8, 9: Hydrolyzable portions of siderophore backbones shown in blue.
Although siderophores are bacterial structures, they’re helpful to humans using the above chemistry to chelate iron and either acquire the metal or sequester it away. Medicinally, siderophores act as sneaky antibiotics through a “Trojan-Horse” strategy. These drugs kill toxic bacterial cells by covalently linking an antibiotic agent to the backbone of a siderophore. The modified siderophore still binds to Fe(III) and the bacterial cell–which is unable to sense the new cytotoxic agent–allows the complex inside.10 Siderophores also help with iron excess diseases (hemochromatosis, iron poisoning) and malaria by chelating Fe(III). In the case of the former, this helps remove iron buildup; in the latter, Fe(III) is sequestered away from cells inside the parasite (which leads to death).5
Sources:
1Hider, R.C., Bickar, D., Morrison, I.E.G., Silver, J. Siderophore iron-release mechanisms. J. Am. Chem Soc., 1984, 106: 6983-6987.
2Górska, A., Sloderbach, A., Marszałł, M.P. Siderophore–drug complexes: potential medicinal applications of the ‘Trojan horse’ strategy. Trends in Pharm. Sci. 2014, 35: 442-449.
3Dhungana, S., Crumbliss, A. L. Coordination chemistry and redox processes in siderophore-mediated iron transport. Geomicrobiol. J. 2005, 22: 87-98.
4Stintzi, A., Barnes, C., Xu, J., Raymond, K. N. Microbial iron transport via a siderophore shuttle: A membrane ion transport paradigm. PNAS. 2000, 97: 10691-10696.
5Ali, S. S.; Vidhale, N. N. Bacterial siderophore and their application: A review. Int. J. Curr. Microbiol. App. Sci 2013, 2, 303–312.
6Hider, R.C., Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27: 637-657.
7Image of 1BY5 (Locher, K.P., Rees, B., Koebnik, R., Mitschler, A., Moulinier, L., Rosenbusch, J.P., Moras, D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998, 95: 771-778) created with Chimera (Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25: 1605-1612).
8Image of 2XUZ (Peuckert, F., Ramos-Vega, A.L., Miethke, M., Schwoerer, C.J., Albrecht, A.G., Oberthuer, M., Marahiel, M.A. The siderophore binding protein FeuA shows limited promiscuity toward exogenous triscatecholates. Chem. Biol. 2011, 18: 907-191) created with Chimera (Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25: 1605-1612).
9Image of 3MWF (Grigg, J.C., Cheung, J., Heinricks, D.E., Murphy, M.E. Staphylococcus aureus SirA specificity for staphyloferrin B is driven by localized conformational change) created with Chimera (Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25: 1605-1612).
10Miethke, M., Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Molec. Biol. Rev. 2007, 71: 413-451.
11Ahmed, E. Holmström, S.J.M. Minireview: Siderophores in environmental research: roles and applications. Microbial Biotech. 2014, 7: 196-208.
12Eisenhauer, H.A., Shames, S., Pawelek, P.D., Coulton, J.W. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J. Biol. Chem. 2005, 280: 30574-30580.
13Bertini, I., Gray, H.B., Stiefel, E.I., Valentine, J.S. (2006). Siderophores. In Stiefel, J. (Ed.), Biological inorganic chemistry: Structure and reactivity (151-160). Sausalito, California: University Science Books.
14Peuckert, F., Ramos-Vega, A.L., Miethke, M., Schwoerer, C.J., Albrecht, A.G., Oberthuer, M., Marahiel, M.A. The siderophore binding protein FeuA shows limited promiscuity toward exogenous triscatecholates. Chem. Biol. 2011, 18: 907-191
15Grigg, J.C., Cheung, J., Heinricks, D.E., Murphy, M.E. Specificity of Staphyloferrin B recognition by the SirA receptor from Staphylococcus aureus. J. Biol. Chem. 2010, 285: 34579-34588.
16Cooper, S.R., McArdle, J.V., Raymond, K.N. Siderophore electrochemistry: Relation to intracellular iron release mechanism. Proc. Natl. Acad. Sci. USA. 1978, 75: 3551-3554.
17Lippard, S.J., Berg, J.M. (1994). Bioavailability of metal ions. In Kelly, A. (Ed.), Principles of Bioinorganic Chemistry (105-139). Mill Valley, California: University Science Books.
Contributed By:
This work was originally written by Emma Bridgman, Fall 2016: Emma is currently (as of 2016) a senior chemistry major at Saint Mary's College in Notre Dame, IN.
This work was originally edited by Dr. Kathryn Haas, Assistant Professor at Saint Mary's College.


