Iron Superoxide Dismutase (FeSOD)
- Page ID
- 98120
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)FeSOD
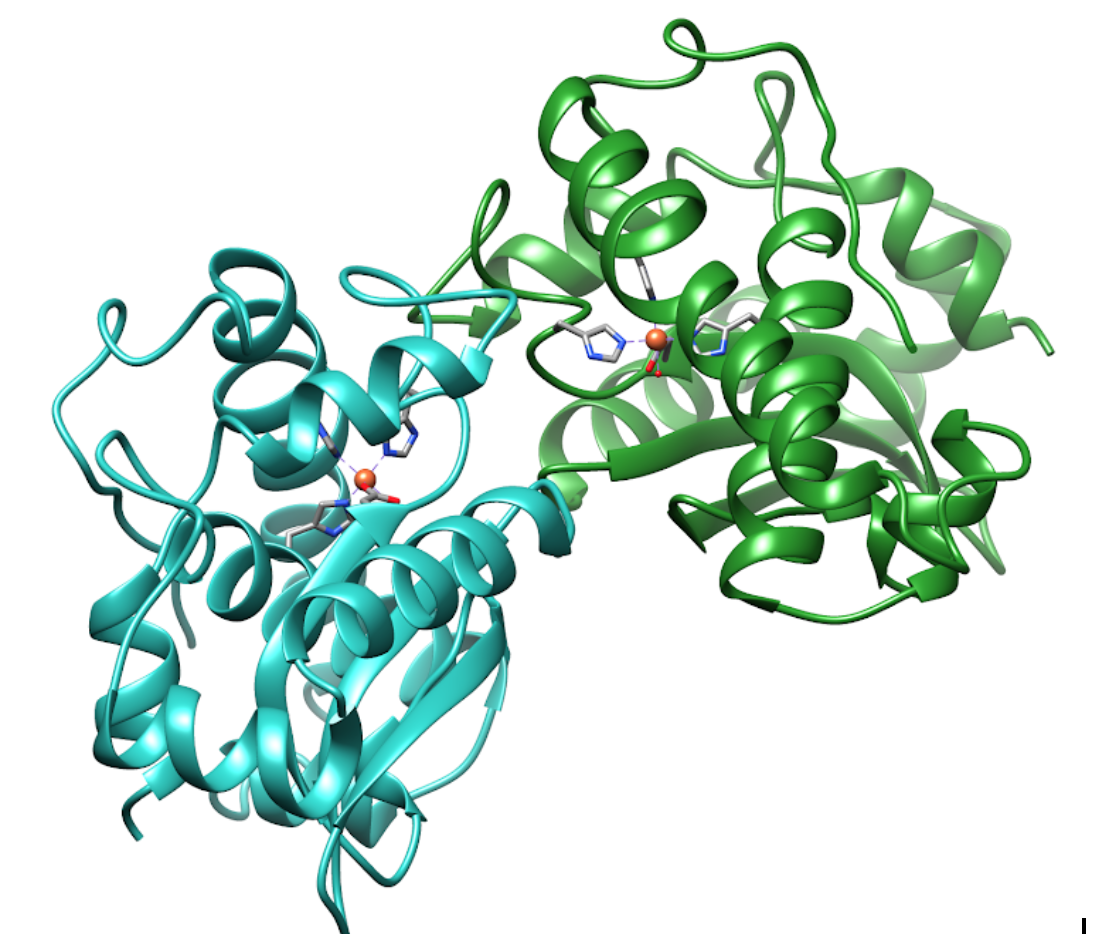
Figure 1: The Iron Superoxide Dismutase Molecule in 3-dimensions.
Iron Superoxide dismutase (FeSOD) is a member of a larger class of superoxide dismutase (SOD) proteins. SOD proteins convert superoxide(O2-) into hydrogen peroxide (H2O2) and dioxygen (O2). These enzymes work by reducing and oxidizing superoxide through a reaction called disproportionation (1).
Disproportionation of superoxide: 2O2- + 2H+ → O2 + H2O2 (1)
Superoxide ( O2-) is a radical species formed during normal aerobic metabolism. It can be toxic in some systems but is also involved in chemical signaling. Because O2- can be toxic, the dismutation reaction catalyzed by SOD is essential to life. SOD helps to reduce oxidative stress caused by aerobic metabolism within the generator of the cell, the mitochondria. Reactive oxygen species (ROS), like superoxide, have been shown to cause cell damage. Antioxidant catalysts, like SODs, help to regulate and “deactivate” ROS by converting them to less harmful molecules, like dioxide (O2) and hydrogen peroxide (H2O2), as shown in reaction (1).
FeSOD is thought to be the first SOD to evolve due to the abundance of iron and low levels of oxygen in Earth’s primitive atmosphere.1 It was first discovered in 1973 in yeast (E. coli) by Yost et. al1 , and has since been found in archaea, bacteria, protists, and some primitive plants within the cytosol, glycosomes, and mitochondria. Over time, Earth’s atmosphere gave way to higher concentrations of oxygen gas, which reacted with iron to make insoluble iron oxides (rust). The decrease in iron solubility made iron less biologically-available to organisms and promoted life to develop SODs that use other, more abundant metals like copper, zinc, and nickel.1
FeSOD is a soluble globular protein with two mirrored domains. Each domain contains an Fe(III)-coordinating activate site
During the dismutation reaction, the iron ion at the active site cycles between Fe(III) with five d-electrons, and Fe(II), with 6 d-electrons. Figure 2 depicts the active site for FeSOD. It is clear from this figure that the ligand configuration around Fe(III) involves three His groups, one Asp group, and one water molecule in a slightly distorted from trigonal bipyramidal configuration.3
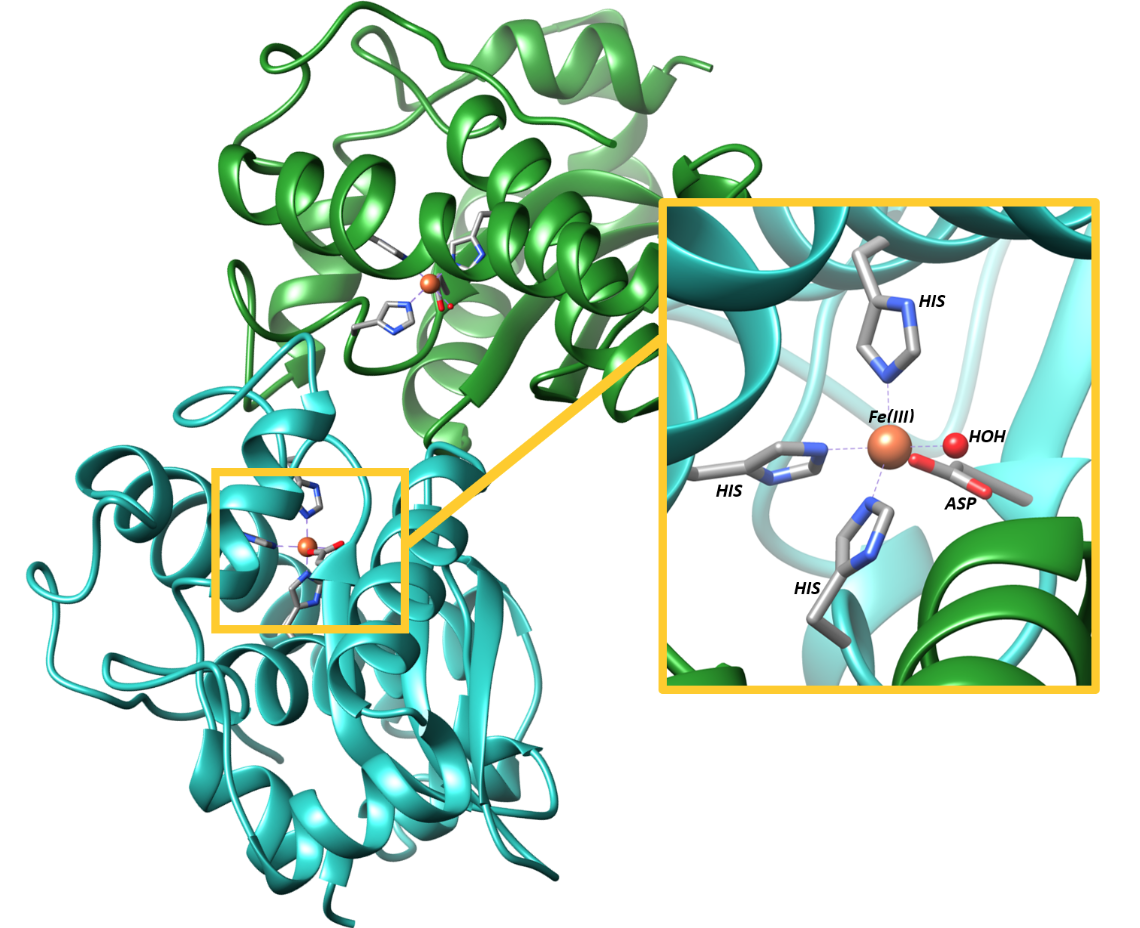
Figure 2: The active site for FeSOD is characterized by two equatorial Histidines, an axial Histidine, an equatorial Aspartic Acid, and an axial water coordinated to the iron in a trigonal bipyramidal geometry. The fourth nitrogen of each His group, the oxygen of the water, and the negative oxygen of the Asp are the specific donor atoms1,2
The differentiator of FeSOD from later-evolved SOD’s is the use of Fe as the metal cofactor rather than more biologically abundant metals. Despite the low solubility of Fe and the challenges of Fe acquisition, FeSOD binds selectively and strongly to Fe.
The ability of FeSOD to retain Fe in biological solutions can be explained by its high affinity. This can be attributed to the stability created from the chelate effect. A chelator is any ligand that binds to a metal ion with more than one donor atom. In the case of FeSOD, there are multiple donor groups to the iron ions thus classifying FeSOD as a chelator. To be specific, there are four donor groups for FeSOD; three histidines (His) and one aspartic acid (Asp). Thus FeSOD is a tetradentate ligand. The benefit of having several ligands all binding to the iron ion is that it will stabilize the ion through an increase in the entropy of the system. 4 In general, the higher the number of ligands binding to the metal, the higher the entropy, and the more stable the ion will be.
One other cause for the affinity and selectivity of FeSOD can be explained through hard soft acid-base theory (HSAB) and the fact that the iron ions are Lewis acids. In HSAB theory, hard acids/bases are characterized by the ratio of charge to ionic size (charge density). A hard acid or base is an atom or ion whose charge is large in relation to its radius. Fe(III) is a Lewis acid with, a high charge to radius ratio, so it is classified as a hard acid. The nitrogens of the three His groups in the active site of FeSOD are borderline bases, while the oxygen of the Asp group is a hard base. Likewise, the Fe(III) and Fe(II) ions are hard and borderline, respectively. According to HSAB, like will bind to like, so the His and Asp groups will prefer hard or borderline acids to softer ones. It is worth noting, however, that FeSOD has been shown to also bind to Manganese, though this substitution won’t convert superoxide. Mn(III) is considered a hard acid similar to Fe(III), so this is a logical switch.
FeSOD is not octahedral like many metal-ligand systems and instead takes on a distorted trigonal bipyramidal geometry as shown above.3 Ligand Field Theory (LFT) is the best explanation for this geometry. According to LFT and crystal field theory (CFT), the increase in formal charge will draw the ligand electrons closer to the metal ion. This results in higher electron-electron repulsion and a splitting of the d-orbitals as well as an increase in the energy between bonding and antibonding orbitals (Δ). The Δ is significant because it relates to how the ligands interact with the metal. The greater the Δ, the more unpaired electrons there will be; thus, even though both ions are considered high spin in relation to other metals, Fe(III) will have fewer electron pairs than Fe(II) (Figure 3). High and low spin refers to the number of unpaired electrons, and the high spin of Fe can be attributed to its formal charge of 2+/3+ as well as its position as a 3d6/5 metal. Additionally, the small Δ is logical because the donor atoms in this active site are both σ-donating and π-accepting. The donating or accepting character of the ligands has to do with the electron configurations of the donor atoms and when a donor atom is π-accepting, the Δ will be lowered. For the His groups, the nitrogen will be σ-donating and π-accepting because the nitrogen has an available pair of electrons which can coordinate to the Fe ion as well as an empty π* orbital; likewise, the Asp and water will be σ-donating because the oxygen atoms have an available pair of electrons but no empty π* orbital.
These electronic characteristics of a metal-ligand complex can be experimentally observed using UV-Vis spectroscopy. The excitation and relaxation of electrons release photons of light which can be detected using a UV-Vis Spectrophotometer. The intensity of these readings directly relates to the Δ value, and the absorbance (color) relates to the energy of the complex, and thus the spin.4 For example, a complex which appears yellow absorbs violet light, so the light high energy; thus, the ligands will be strong field, have low spin, and a large Δ.4 Experimental analysis of FeSOD has been accomplished using UV-Vis Spectroscopy.3 The resulting spectra can be attributed to the ability of Fe(III) and Fe(II) to do d-d-electron transfer as well as metal-ligand charge transfer (MLCT) (Figure 3).4 Fe(III) is d6 metal ion, so it has 4 unpaired electrons (low spin) or two unoccupied d-orbitals (high spin), leaving plenty of room for excitation of electrons into higher d-orbitals. The same is true for Fe(II) as a d5 metal ion. Additionally, MLCT is possible because of the π-accepting nature of the ligands.
One UV-Vis spectral study of FeSOD analyzed its activity in cyanobacterial cells. In these experiments, FeSOD was observed at 280 nm and 350 nm, and the extinction coefficient was reported as ϵ280 nm= 46410 M-1cm-1 and ϵ350 nm= 1850 M-1cm-1. The first peak at 280 nm represents the amino acids that have aromatic groups, i.e. groups which are spectroscopically active such as Tyr. The peak at 350 nm is indicative of the Fe(III) in FeSOD and as the ion is reduced to Fe(II), the peak absorbance will decrease.5 Based on the high (ϵ>> 1000) extinction coefficient, metal-ligand charge transfer (MLCT) is likely the major cause for this UV-Vis absorption.
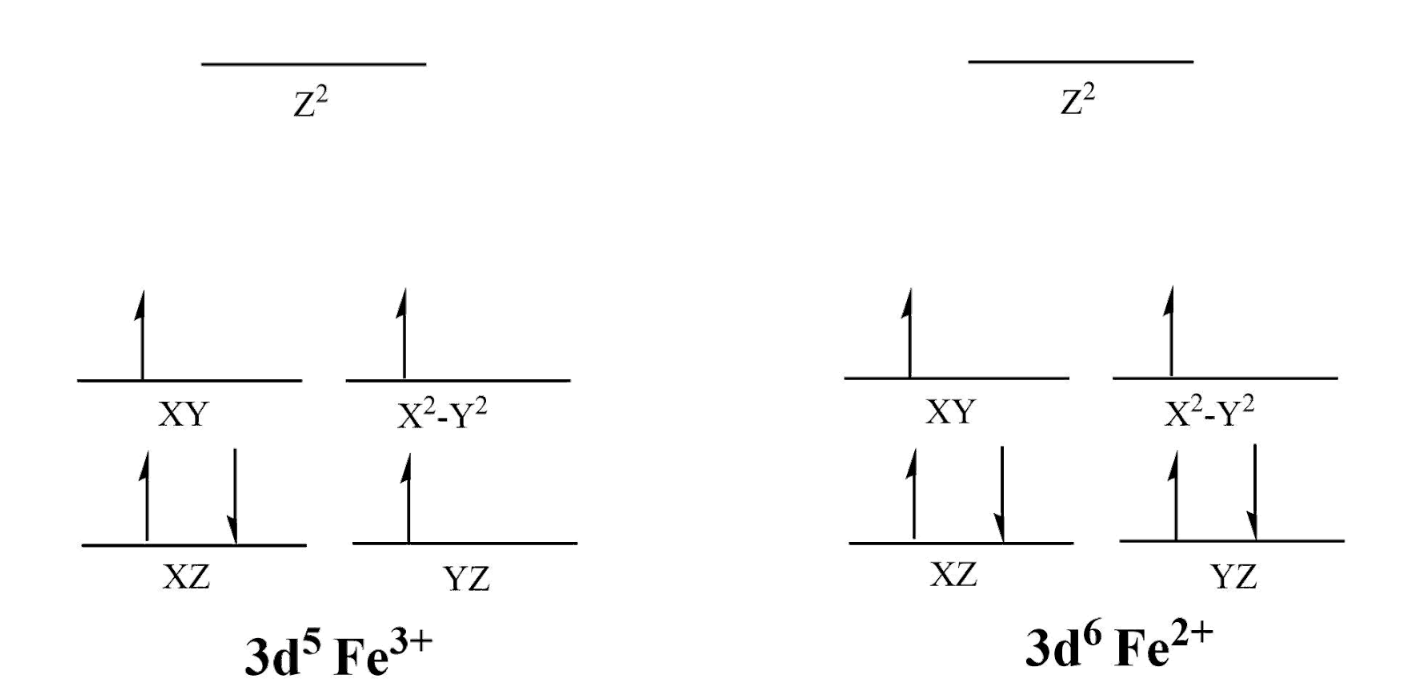
Figure 3: The splitting diagrams for a 3d5 Fe(III) and 3d6 Fe(II) trigonal bipyramidal complexes. It is clear that these metals will be high spin because of their unpaired electrons. Additionally, the stability of the metal complex increases as Fe(III) is reduced.
The Jahn-Teller distorted geometry also plays a pivotal role in the electronic behavior and catalytic mechanism of FeSOD. The slight distortion means that Fe(III) will not be as stable as it would be in a normal trigonal planar geometry, providing the opportunity for reduction.8,5 Instead of a rigid metal center, the d5 Fe(III) has ability to add an electron from the superoxide to form a more stable d6 Fe(II) (See Figure 3).Fe(III) will coordinate with the superoxide ion and undergo a process called Inner-Sphere Electron Transfer.1 Inner sphere electron transfer implies the superoxide will coordinate to the iron ion while bound to the active site of FeSOD and hydrogen bond to neighboring residues, then transfer an electron to the Fe(III). This results in the donation of the radical electron from superoxide to the iron, thus reducing iron. During this process, Fe(III) is reduced by the superoxide to Fe(II) through a REDOX reaction.2,1 Fe+3 has a reduction potential (E0’) of 0.77V, and the reduction potential for FeSOD is reported to be 0.1V.1 These reduction potentials are important to note because they give us insight into the favorability of this reaction. The positive nature of the Fe(III) reduction potential identifies Fe(III) as the target for reduction. Similarly, the positive nature of the overall E0’ for FeSOD demonstrates that this will be a favorable, spontaneous reaction. The half reactions of the catalytic mechanism of FeSOD are as follows:
O2- + H++HO--Fe+3SOD → O2 + H2O- Fe+2SOD (2)
O2- + H++H2O-Fe+2SOD → H2O2 +HO- - Fe+3SOD (3)
In this mechanism, the reduction of Fe(III) creates a proton gradient which supplies the necessary protons, via hydrogen bonding, for neutralizing the superoxide radical.1 The reaction mechanism for FeSOD is described in Figure 4.

Figure 4: The mechanism for FeSOD.1
From this catalytic mechanism, it is clear to see that superoxide contributes to the reaction. Between step 1 and 2 is the inner-sphere REDOX described earlier. The superoxide radical bonds to Fe(III) as well as the hydrogen on the neighboring Tyr, while donating its radical electron to Fe(III). This step effectively reduces Fe(III) to Fe(II) and releases oxygen gas. In step three, superoxide makes another appearance but instead of coordinating to iron, it binds to the hydrogen atom of Tyr, as before, and of water. Superoxide effectively pulls these hydrogens closer to itself, creating a positive charge on the oxygen in water. This change creates the opportunity for more inner sphere REDOX as the iron donates an electron to the oxygen, making it negative (step 4). Now the iron ion is back to its original, and more stable state, and the oxygen in water is negative. This negative charge leads to the water donating one of its hydrogens to superoxide, which in the presence of hydrogen ions become hydrogen peroxide and the cycle begins again.1 Additionally, this inclusion of positive hydrogen ions suggests that the pH of the solution surrounding FeSOD could hinder the catalytic function if it is too basic.
Since FeSOD helps to catalyze the reduction of O2- to O2 using metal-ligand interactions with Fe(III), the electron count of the metal complex likely affects the activity of FeSOD. Each intermediate has a separate electron count. The first intermediate will have 15 electrons, two electrons for each coordination to a ligand, and five d-electrons in Fe(III). This process is similar for the rest of the intermediates, and the electron counts are labeled in Figure 5.
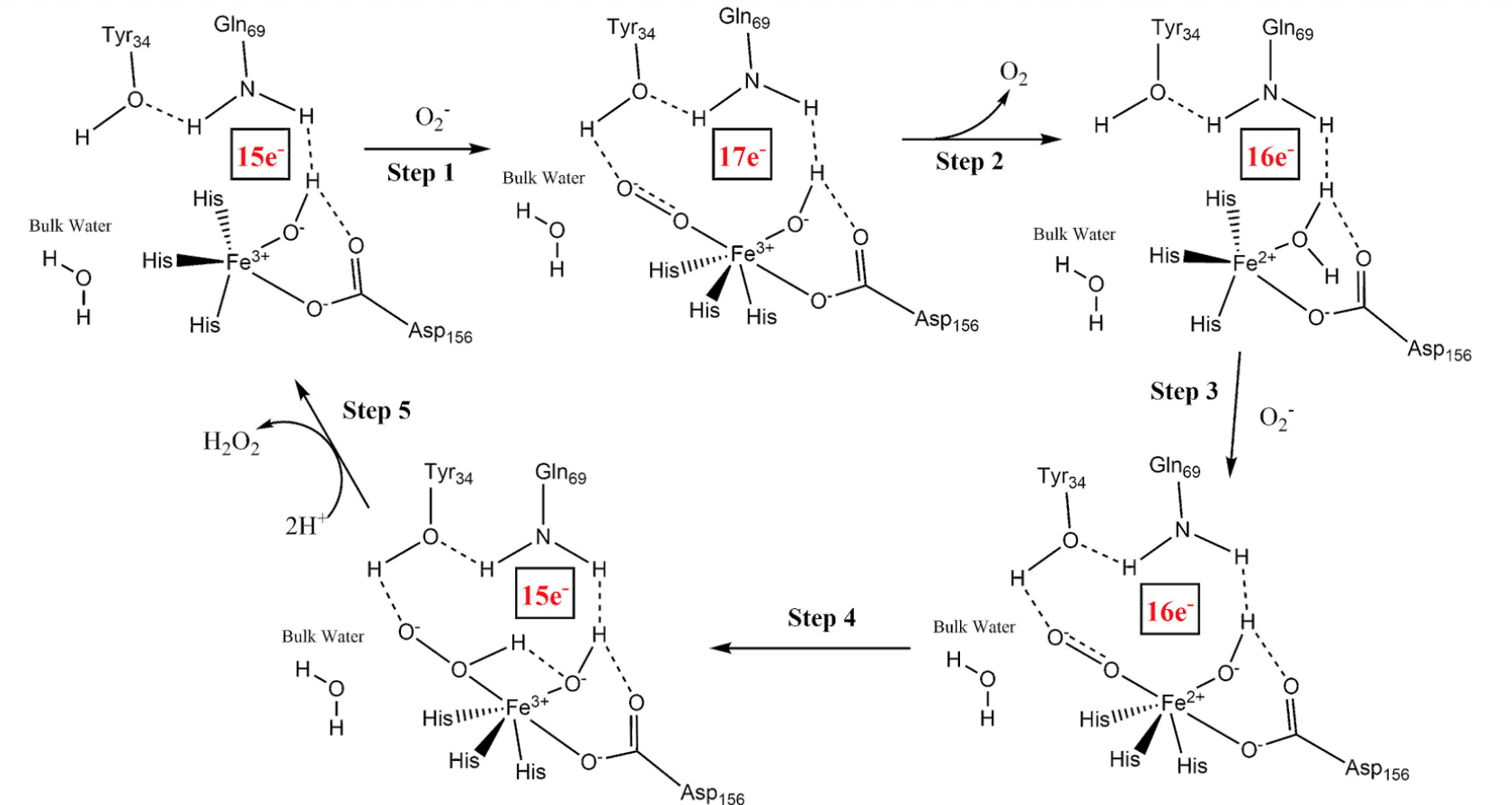
Figure 5: The mechanism for FeSOD with electron counts (in red).1 As the electron count increases the stability of the complex increases; however FeSOD functions because of its slight instability. So even though there is an increased stability when superoxide
Knowing the electron count is helpful because it explains the stability of the iron throughout the mechanism. For example, the lower electron counts, 15 electrons, suggest lower stability, such as the last and first intermediates. This pattern of an increasing and then decreasing electron count makes sense because it shows that the mechanism creates favorable stability for FeSOD while still converting superoxide into oxygen gas and dihydrogen peroxide.7 Without this stability, the catalytic cycle may not proceed. The similarity in stability between intermediates is vital to the thermodynamic progression because large changes in stability mean higher changes in energy from step to step. Lowering the change in energy between intermediates means the reaction will produce less entropy.
FeSOD and other SOD molecules are vital to life on Earth because of their ability to remove ROS, and as the environment around us continues to change as evolution progresses, SODs will adapt to fit our needs. These amazing molecules, specifically FeSOD, have been explained through theories and different experiments, but many still questions remain. With dedicated research, more will be discovered about how FeSOD functions, how organisms who use it have evolved, and how its existence contributes to life.
Sources
- Sheng, Y.; Abreu, I. A.; Cabelli, D. E.; Maroney, M. J.; Miller, A.-F.; Teixeira, M.; Valentine, J. S. Superoxide Dismutases and Superoxide Reductases. Chem. Rev. 2014, 114 (7), 3854–3918
- Bertini, I.; Gray, H. B.; Stiefel, E. J.; Valentine, J. S. Biological Inorganic Chemistry: Structure and Reactivity; University Science Books: Sausalito, Californie, 2007.
- Jean Philippe Renault; Catherine Verchère-Béaur, and; Morgenstern-Badarau*, I.; Yamakura, F.; Gerloch*, M. EPR and Ligand Field Studies of Iron Superoxide Dismutases and Iron-Substituted Manganese Superoxide Dismutases: Relationships between Electronic Structure of the Active Site and Activity http://pubs.acs.org/doi/abs/10.1021/ic0000451 (accessed Feb 9, 2018).
- Chelation https://chem.libretexts.org/Core/Inorganic_Chemistry/Coordination_Chemistry/Complex_Ion_Equilibria/Chelation (accessed Feb 8, 2018).
- Haas, K; Metal Ligand Interactions Review Material; 2018
- Hard and Soft Acids and Bases. (2016, August 21). Retrieved March 7, 2018, from https://chem.libretexts.org/LibreTexts/Saint_Mary’s_College%2C_Notre_Dame%2C_IN/CHEM_342%3A_Bio-inorganic_Chemistry/Chapters/Chapter_2%3A_Introduction_to_Metal-Ligand_Interactions/2.2_Acids_and_Bases/B._Hard_and_Soft_Acids_and_Bases
- Gregory, E. M. Characterization of the O2-Induced Manganese-Containing Superoxide Dismutase from Bacteroides Fragilis. Arch. Biochem. Biophys. 1985, 238 (1), 83–89.
- Introduction to Crystal Field Theory - Chemistry LibreTexts https://chem.libretexts.org/LibreTexts/Saint_Mary’s_College%2C_Notre_Dame%2C_IN/CHEM_342%3A_Bio-inorganic_Chemistry/Chapters/Chapter_3%3A_Introduction_to_Advanced_Bonding_Theories/3.1_Crystal_Field_Theory/A._Introduction_to_Crystal_Field_Theory (accessed Feb 15, 2018)
- Haas, K; Metal REDOX Review Material; 2018
Contributed By:
This work was originally written by Elizabeth Innis, Spring 2018: Elizabeth is currently (as of 2018) a junior chemistry major at Saint Mary's College in Notre Dame, IN.
This work was originally edited by Dr. Kathryn Haas (Assistant Professor), Madison Sendzik (Teaching and Research Assistant), and Dr. Dorothy Feigl (Professor) at Saint Mary's College.

