HIV-1 Nucleocapsid Protein
- Page ID
- 98119
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)What exactly is HIV-1?
Human immunodeficiency virus type 1 (HIV-1) attacks the body’s immune system by targeting specific CD4 cells, or T cells, that help the body fight off infections. Over time, the virus continues to destroy these cells, thus, weakening the body’s immune system. Lack of treatment can reduce the number of these cells significantly, leading to acquired immunodeficiency syndrome (AIDS), which is characterized by the onset of frequent severe illnesses.1
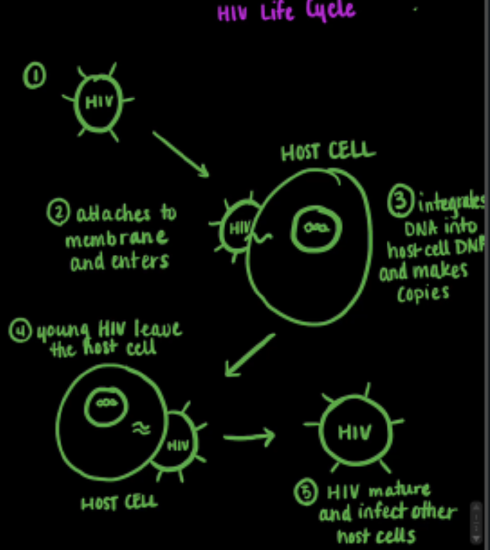
Figure 1. Drawing of the HIV-1 life cycle. The mature HIV-1 retrovirus attaches itself to a host cell, where it integrates its DNA into the host cell’s DNA. Replication of the HIV-1 cell can then occur to produce young HIV-1 cells, which then are able to mature and infect other cells.
HIV-1 is a retrovirus, meaning its genetic material uses RNA instead of DNA. Once the virus enters the cell, it uses an enzyme, reverse transcriptase, to convert the viral RNA to DNA. The viral DNA is then integrated into the host cell’s DNA, allowing the HIV-1 retrovirus to replicate and spread in the human body, shown in Figure 1 above.2 The HIV-1 genome is comprised of nine genes, all necessary for the formation of an infectious cell. One of the most important genes is the group-specific antigen, or gag gene, which has the genetic material necessary for the coding of core structural proteins in the retrovirus. The gag gene alone is able to synthesize, transport to the plasma membrane, and assemble the structural precursor polyprotein Gag, shown in Figure 2 below.2 This polyprotein is important because many of the structural proteins needed to produce a mature and infectious HIV-1 particle are derived from this polyprotein. This page focuses on one of these structural proteins, the HIV-1 nucleocapsid protein.2
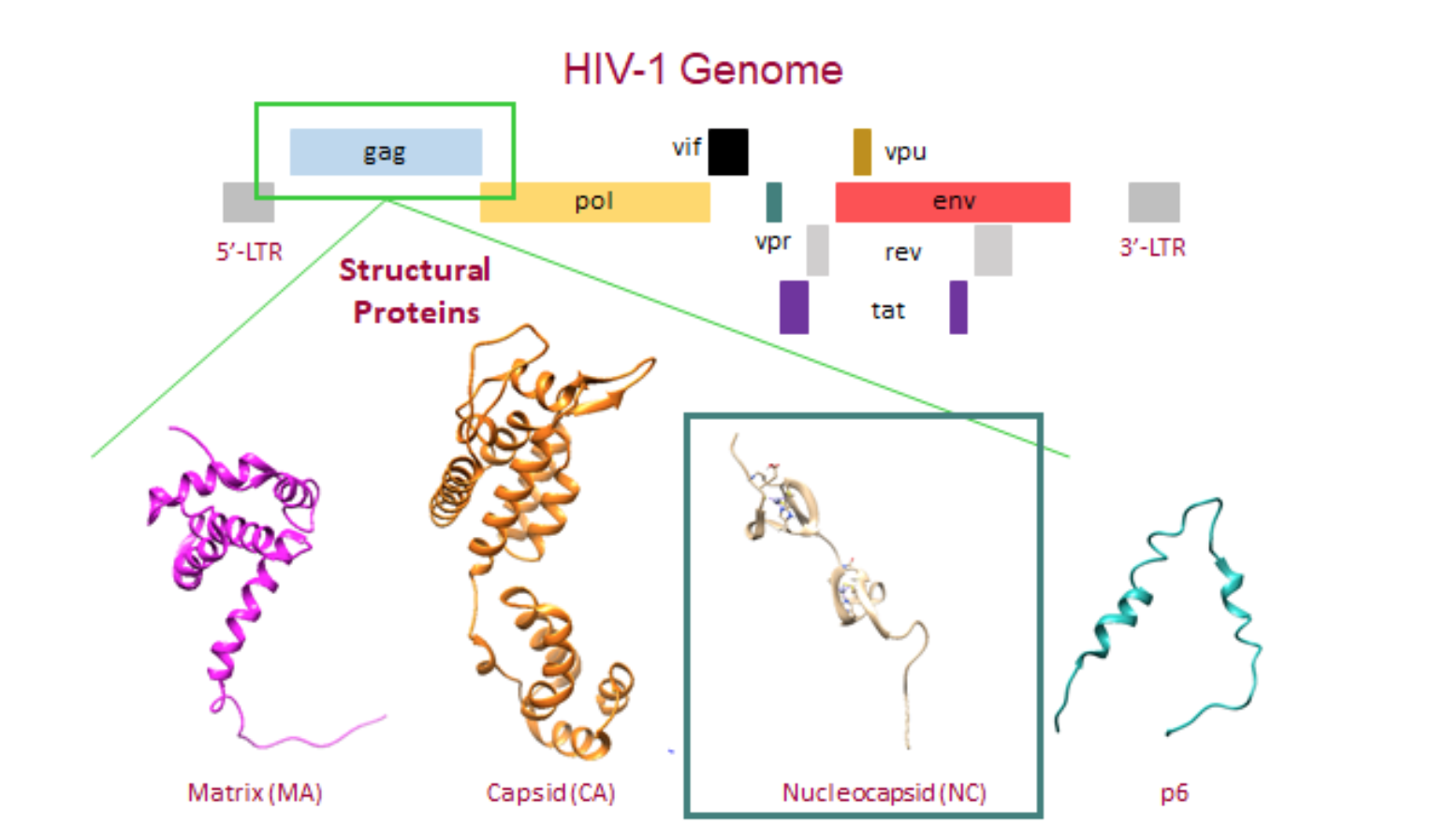
Figure 2. The HIV-1 genome and the structural proteins derived from the gag gene. The gag gene allows for the assembly of the Gag polyprotein which is comprised of the matrix (MA) (PDB: 2H3F) , capsid (CA) (PDB: 3GV2), nucleocapsid (NC) (PDB: 1MFS), and p6 (PDB: 2C55) proteins.
The HIV-1 nucleocapsid protein (NC) is a small, basic protein that acts as a nucleic acid chaperone.3 The NC plays a hand in multiple functions of the virus replication cycle, including RNA packaging and virus assembly, reverse transcriptase, and replication.4 As a nucleic acid chaperone, the NC protein has the ability to remodel nucleic acid structures to their most thermodynamically stable conformation. This ability plays a fundamental role in the HIV-1 replication and reverse transcriptase processes.3 Figure 3 below shows the NC protein, which contains a highly basic N-terminus, a short C terminus, and two zinc binding domains, also known as a zinc (Zn) finger, connected by a short peptide linker.3
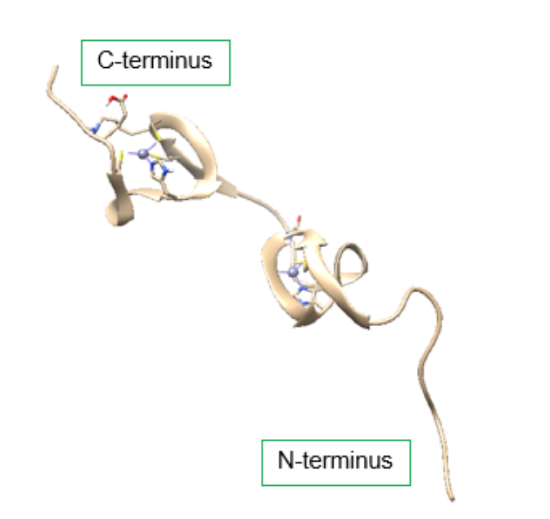
Figure 3. Structure of the HIV-1 nucleocapsid protein (PDB: 1MFS). Each terminus contains a zinc finger motif, which contains a Zn2+ ion bound to a ligand containing three cysteine amino acid residues and one histidine amino acid residue. The zinc binding motifs provide stabilization for the small protein. DOI: 10.2210/pdb1MFS/pdb
Very small protein domains often require some sort of structural stabilization, which can often be accomplished through the binding of metal ions. Shown in Figure 4 below, the zinc binding domains are comprised of the metal ion, Zn2+, and a four amino acid ligand in the sequence Cys-Cys-His-Cys (CCHC motif).3 The presence of the cysteines and the histidines adjacent to each other allows them to grab a Zn2+ ion from the surrounding environment, and fold tightly around that ion.5
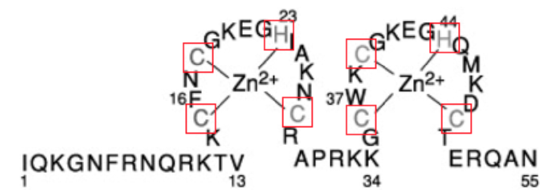
Figure 4. Primary sequence of HIV-1 NC. The cysteine and histidine residues that chelate zinc are shown in gray. The numbers 16 and 37, highlight the hydrophobic residues for each zinc finger, that are utilized for RNA binding. Adapted from (Levin, 2005).4
Each Zn finger contains an aromatic, hydrophobic residue: this is phenylalanine-16 (16F) in the N-terminal Zn finger and tryptophan -37 (37W) in the C-terminal Zn finger, shown in Figure 2. In a mature HIV-1 retrovirus, the aromatic residues from the two Zn fingers are found side by side, as a result of the the flexibility of the peptide linker.6 The closeness of the aromatic, hydrophobic residues creates a hydrophobic patch. This hydrophobic patch provides an ideal platform for RNA binding through stacking interactions with residues in RNA nucleic acid sequences, shown in FIgure 5 below.4 This ideal binding platform acts as a major contributor to HIV-1 NC and RNA nucleic acid interactions.
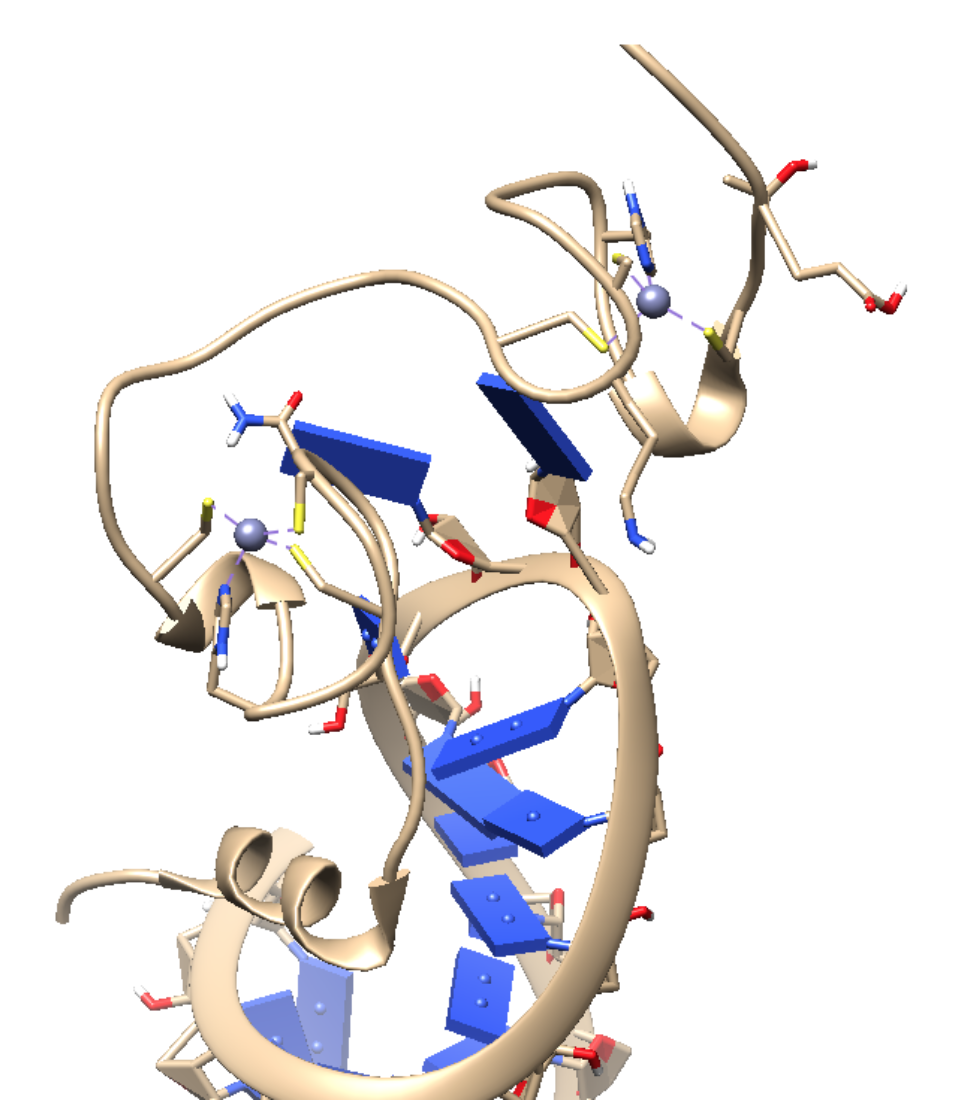
Figure 5. The HIV-1 NC bound to SL3 stem-loop recognition element of the psi-RNA packaging signal (PDB: 1A1T). The recognition of the HIV-1 genome occurs through interactions between the NC and a region of the viral RNA, know as the psi site. This psi site has four different stem loops (SL1-SL4). The SL3 loop is of particular interest because it is highly conserved among different HIV-1 strains, and because linkage of the SL3 stem loop to heterologous RNA allows for recognition and packaging of the RNA.7 DOI: 10.2210/pdb1A1T/pdb
Zn coordination chemistry
Each zinc-finger domain is a tetradentate ligand, binding to Zn with three cysteine sulfur atoms, and one histidine imidazole nitrogen. The bonding interactions between the protein and Zn can be most simply described as an acid/base or donor/acceptor interaction where the cysteines and histidine are acting as Lewis bases (electron pair donors) and Zn acts as a Lewis acid (electron pair acceptor). The interaction between each Zn finger domain and its Zn metal ion is particularly stable, with affinity of approximately [can you find affinities anywhere?], and it is selective for Zn over other biological metals. The stability and selectivity of these interactions can be explained by several principles of inorganic chemistry, described below.
The Chelate Effect:
The three cysteines and one histidine residue provide four coordinating ligands for Zn and they are also connected by the protein backbone (or by peptide bonds). This makes the Zn finger domain a multi-dentate ligand. A multidentate ligand, or in other words a chelator, is a ligand that binds to a metal ion through more than one donor group. As one multidentate ligand, the protein is able to bind to the Zn2+ metal ion with four donor atoms, one from each side chain in the CCHC chelator. This leads to a tetradentate complex that is more thermodynamically stable than an analogous complex of Zn with individual C,C,C,and H amino acids. The increased stability is due to the chelate effect, which is fundamentally explained by an increase in entropy. To illustrate the chelate effect, imagine a reaction that begins with two molecules on the reactants side, the protein (CCHC) and the hydrated Zn metal. However, upon completion of the reaction, the water molecules attached to the Zn ion are replaced with the chelator, resulting in an increase of total molecules to five, as shown in Figure 6 below. The increase in molecules from reactants to products results in an increase in entropy. This increase in entropy is favorable, and therefore, the Zn-CCHC complex has increased stability due to this chelate effect.
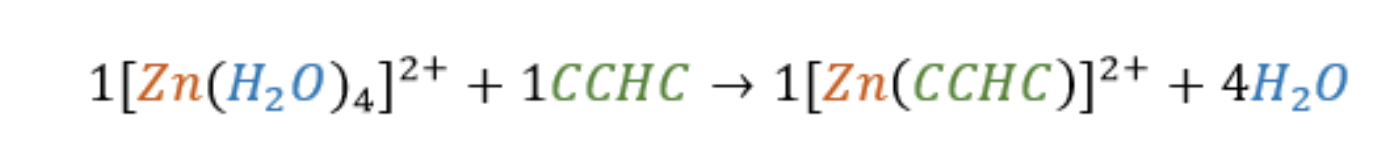
Figure 6. The binding reaction between the CCHC motif and the Zn2+ ion. The numbers show that the reaction encourages an increase in molecules, indicating an increase in entropy. This increase in entropy provides a more thermodynamically stable metal-ligand complex, and therefore, is more favorable.
Hard/Soft Acid Base Theory:
Each of the Zn fingers contain a Zn ion that is bound in a 4-coordinate chelate complex by cysteine thiolate side chains and an imidazole ring found on histidine, as shown in Figure 7 below.6 Both cysteine and histidine are common ligands for Zn2+.8 The appearance of these ligands in a Zn binding site can be rationalized using Hard Soft Acid Base (HSAB) theory. HSAB theory categorizes metals (acids) and ligands (bases)based on “Hard” and “Soft” characteristics. There are three different categories: hard, soft, and borderline. These categories are somewhat dependent on charge density, but can also depend on the type of interaction (covalent vs ionic) that is occurring in the metal-ligand bond. Overall, acids or bases characterized as hard typically have a higher charge density, and those that are soft display a lower charge density. It is important to note that a key idea of HSAB theory is that “like prefers like”. For example hard metals (acids) will tend to prefer binding to hard ligands (bases).
The zinc finger motif on the HIV-1 nucleocapsid protein coordinates Zn2+ via the side chain atoms of three Cys and one His . The zinc ion (Zn2+) is classified as as a borderline acid. The presence of both Cys and His ligands give the HIV-1 metal binding site mixed soft/borderline character due to the presence of the soft thiol group (-S-) on the cysteines and the borderline imidazole nitrogen of the histidine residue. Since “like binds with like”, there is a strong selectivity between the Zn2+ and the cysteine and histidine ligands because of their similar soft acid/base characteristics.
Coordination Geometry: Why tetrahedral?
Ligand Field Theory (LFT), a principle of coordination chemistry, can be applied to explain this troubling question. The full d shell of Zn2+ results in a ligand field stabilization energy (LFSE) of zero. An LFSE of zero comes from a balance between the higher orbital and lower orbital electrons, shown in Figure 8 below. This value indicates that Zn2+ has no LFSE preference for a specific geometry. With no preference for a specific geometry, sterics become the major factor in determining the geometry that Zn2+ takes. Other metal ions, that do not have a full d shell, may have an imbalance between the higher orbital and lower orbital electrons that results in a negative LFSE value. Metal complexes will prefer to adopt the configuration with the lowest energy or most negative LFSE value. This fact indicates that these other metal ions would have a preference for a specific geometry, most likely octahedral. The differences in geometry helps explain the selectivity for the Zn2+ ion over other metals such as iron (Fe) or copper (Cu), which have specific preferences for non-tetrahedral geometries.
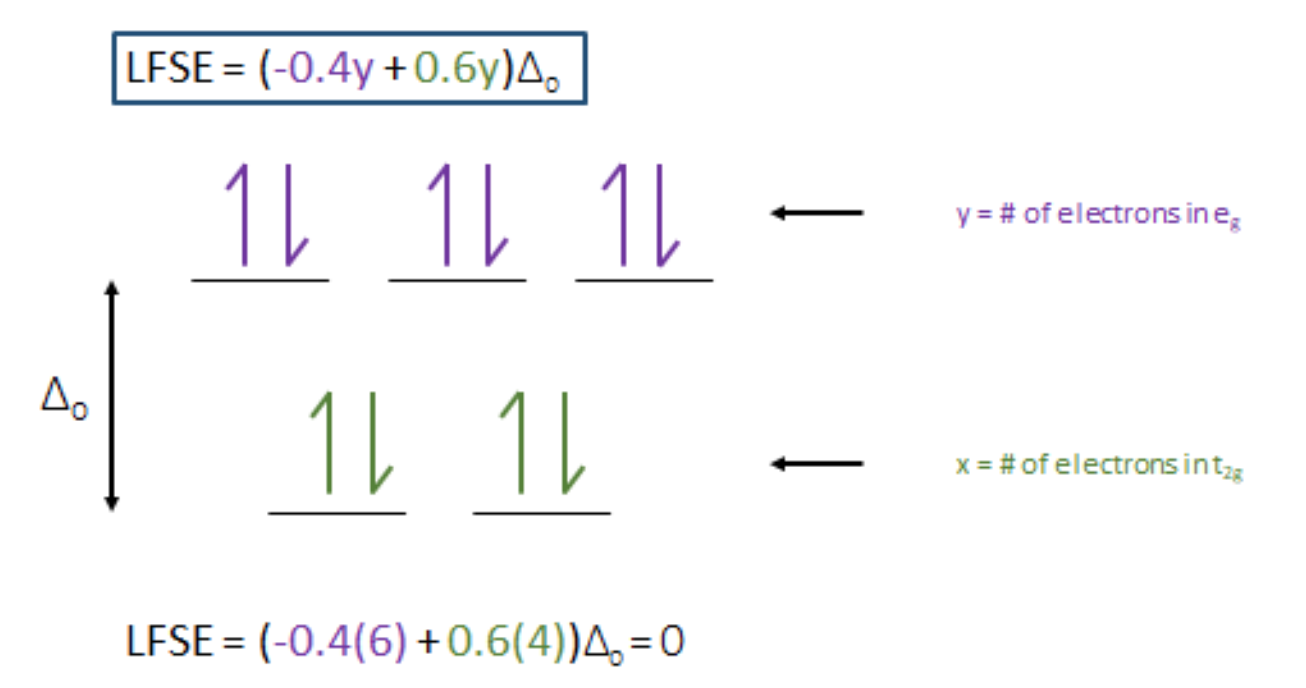
Figure 8. LFSE splitting diagram for Zn2+. An LFSE = 0 indicates that the Zn ion has no LFSE preference for any geometry. With no LFSE preference sterics become the major factor in determining the geometry for Zn finger complex.
As stated above, sterics play a major role in the coordination geometry around the Zn ion. Zn is a relatively small ion, and the more ligands bound to the ion, the more unfavorable steric interactions will occur. Knowing this, we might predict that Zn will favor a 4 coordinate geometry over a 6 coordinate geometry to reduce steric stresses. The tetrahedral geometry provides a nice balance between these steric factors and electrostatic factors, which comes from the ligand electron density that help stabilize the +2 charge on the Zn ion. Sterics are also the reason that the tetrahedral geometry is chosen over the 4-coordinate square planar geometry, shown in Figure 9 below. Larger angles between the ligands (109.5°) in the tetrahedral geometry provide more favorable steric interactions than those encountered in a square planar geometry (90°).
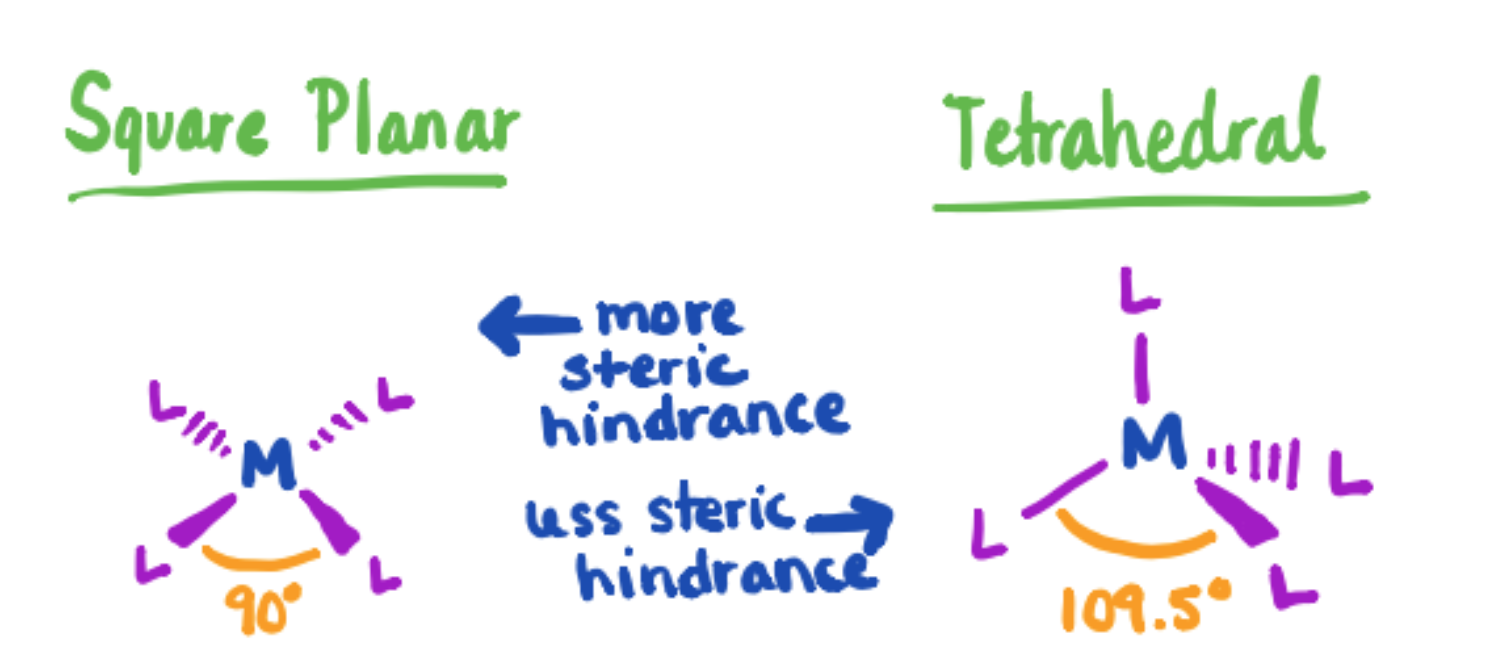
Figure 9. Comparison of square planar and tetrahedral conformations for metal-ligand complexes. The larger angle experienced with the tetrahedral structure provides less steric hindrance. Since sterics are the determining factor in this molecule, a tetrahedral geometry will be more favorable than the square planar geometry.
The 18 electron rule is often apparent in metal catalysts that are known to cycle between 16- and 18- electron species during a catalytic cycle. At an electron count of 18, the metal is said to be in a noble-gas configuration and coordinately saturated, which is highly favorable. Metals that have either lower or higher than 18 electron counts must bond to or lose ligands, respectively. In order to determine the electron count, you must take into account both the number of metal d-electrons and the ligand bonding electrons.
With the HIV-1 nucleocapsid protein, the zinc fingers are bound to the four ligands, resulting in 8 ligand bonding electrons. If we imagine a deconstruction of the complex by separation of the ligands and metal, we achieve a +2 charge on the Zn ion. In the +2 form, we would imagine 2 electrons being removed from the Zn atom to give Zn2+. These two electrons will be removed from the 4s shell because of their lower Zeff due to the extra shielding from the third shell. Under this circumstance,10 electrons remain because Zn has a full d shell. With the addition of the two electrons,a total electron count of 18 is calculated. Through four coordinate bonds with the CCHC motif, Zn is coordinately saturated and stable in a tetrahedral chelate complex.
What are characteristics of tetrahedral geometry?
Molecules that experience a tetrahedral geometry will always be high spin, meaning the splitting energy gap (△) will be small. A small △ means that the lower energy (eg) and higher energy (t2g) orbitals are much closer in energy, allowing for the partial filling of both orbitals before electron pairing. The △ is also influenced by the ligand characterization as a sigma donor only, sigma donor and pi donor, or sigma donor and pi acceptor. For this specific system, the sulfur groups on the cysteine are sigma and pi donors, whereas, the aromatic group on the histidine is a sigma donor and pi acceptor. Ligands that are characterized as sigma and pi donors are considered weak field ligands, and indicated a small △. Since histidine is not a very strong pi-acceptor, the sulfur groups, which are acting as sigma and pi donors, control the system and promote a small △, a characteristic of tetrahedral geometries.
How does LFSE explain kinetics of the molecule?
Typically, the thermodynamics of a molecule can be explained using LFSE. Molecules with lower (more negative) LFSE values will be more thermodynamically stable because of the imbalance between the stabilization of the lower energy orbitals and the destabilization of the higher energy orbitals. Molecules with an LFSE = 0, as mentioned previously, have a balance between these lower and higher energy electrons, and therefore lack structural stability. This allows for the molecule to be thermodynamically unstable and labile, meaning able to react fast.
Within the HIV-1 NC, an LFSE=0 is indicated, as previously mentioned. It is expected that the molecule would be labile and thermodynamically unstable because of its LFSE value of zero. However, within this molecule it is understood that the +2 charge of the Zn2+ metal ion allows for strong electrostatic interactions with the negatively charged ligands resulting in the metal complex adopting more inert qualities compared to a Zn2+ metal complex with more neutral ligands.
Summary
The function of the HIV-1 nucleocapsid protein and the overall function of the HIV-1 virus is dependent on many inorganic chemical principles. By utilizing a Zn finger motif, Zn can act as a structural scaffold so that the small protein is able to a well-defined structure and provide an ideal surface for RNA binding. The structure is also more stable as a result of the chelate effect, where an increase in entropy is seen as the hydrated Zn releases its water molecules and binds to the CCHC multidentate ligand. According to the HSAB theory, the Zn finger is able to make stronger bonds with the cysteine and histidine residues of the CCHC motif because of the similar “soft” characteristics that this specific metal and ligands share. Ligand Field theory shows that Zn2+ has no LFSE preference for any specific geometry, thus the tetrahedral geometry that Zn2+ experiences is determined primarily by sterics. This specific geometry explains the selectivity for Zn over other metal ions that prefer non-tetrahedral geometries. With an LFSE of zero, it would be expected that the molecule would be labile. However, electrostatic interactions between the positive charge of the Zn2+ ion and the negatively charged ligands results in more inert capabilities.
Sources
[1] What are HIV and AIDS? HIV.gov https://www.hiv.gov/hiv-basics/overv...e-hiv-and-aids (accessed 3/15/18)
[2] Scarlata, S.; Carter. C. Role of HIV-1 Gag domains in viral assembly. BBA - Biomembranes 2003, 1614, 62-72
[3] Levin, J. G.; Mitra, M.; Mascarenhas, A.; Musier-Forsyth, K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010, 7, 754-774
[4] Levin, J. G.; Guo, J.; Rouzina, I.; Musier‐Forsyth, K. Nucleic Acid Chaperone Activity of HIV‐1 Nucleocapsid Protein: Critical Role in Reverse Transcription and Molecular Mechanism. In Progress in Nucleic Acid Research and Molecular Biology; Elsevier, 2005; Vol. 80, pp 217–286.
[5] Darlix, J.-L.; Godet, J.; Ivanyi-Nagy, R.; Fossé, P.; Mauffret, O.; Mély, Y. Flexible Nature and Specific Functions of the HIV-1 Nucleocapsid Protein. Journal of Molecular Biology 2011, 410 (4), 565–581.
[6] Aduri, R.; Briggs, K. T.; Gorelick, R. J.; Marino, J. P. Molecular Determinants of HIV-1 NCp7 Chaperone Activity in Maturation of the HIV-1 Dimerization Initiation Site. Nucleic Acids Research 2013, 41 (4), 2565–2580.
[7] De Guzman, R. N.; Rhong Wu, Z.; Stalling, C. C.; Pappalardo, L.; Borer, P. N.; Summers, M.F. Structure of the HIV-1 Nucleocapsid Protein Bound to the SL3 psi-RNA Recognition Element. Science 1998, 279, 384-388
[8] Pace, N. J.; Weerapana, E. Zinc-Binding Cysteines: Diverse Functions and Structural Motifs. Biomolecules 2014, 4, 419-434
Contributed by:
This work was originally written by Emily Najacht, Spring 2018: Emily is (as of 2018) a recent Chemistry graduate of Saint Mary's College. She will graduate with her dual degree in Environmental Engineering from the University of Notre Dame next Spring.
This work was originally edited by Dr. Kathryn Haas (Assistant Professor), Madison Sendzik (Teaching and Research Assistant), and Dr. Dorothy Feigl (Professor) at Saint Mary's College.

