Dopamine Transporter Protein (DTP)
- Page ID
- 97485
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)This is a video introduction to the Dopamine Transporter Protein (PDB 4xp1, 4m48).
Dopamine Transporter Protein (DTP)
Dopamine regulation is a factor in the onset and progression of many mental illnesses. A lack of of dopamine is associated with conditions like anxiety, depression, and Parkinson’s disease. Conversely, a buildup of dopamine leads to euphoric effects felt by recreational drug users.1 The Dopamine Transporter Protein (DAT) is partially responsible for regulating dopamine levels through the reuptake of dopamine in neuronal cells.1 Dopamine is produced in the presynaptic neuron and released to the synaptic cleft, where it may bind to dopamine receptors on the postsynaptic neuron. DAT prevents dopamine accumulation by clearing away unbound dopamine molecules from the synaptic cleft, as shown in the figure below.2
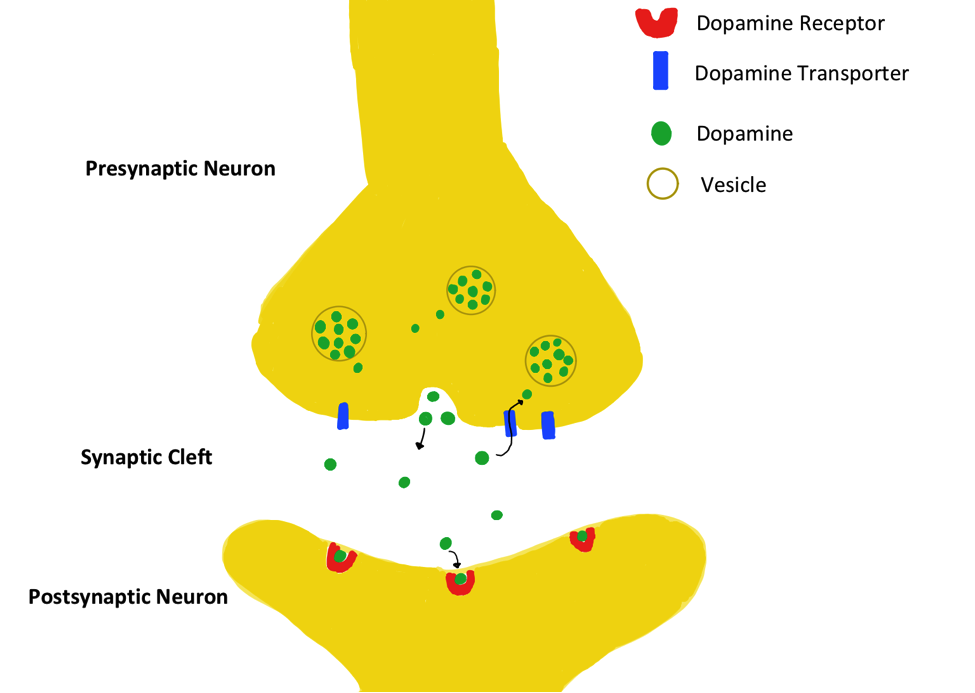
Mechanism of Transport
Dopamine is transported across the membrane from the relatively low-concentration synaptic cleft into the relatively high-concentration presynaptic neuron. This is an energetically expensive process, because the dopamine molecules must be transported against their concentration gradient.3 Rather than using energy from ATP to power this active transport, DAT takes advantage of secondary active transport (also known as ion-coupled transport). The cotransport of one chloride ion and two sodium ions provides the energy to transport the dopamine. The Na+ and Cl- move along their concentration gradient (high to low concentration), allowing the dopamine to move against its concentration gradient (from low to high concentration) by making the net reaction energetically favorable.3 This cotransport is demonstrated in figure 2 below.
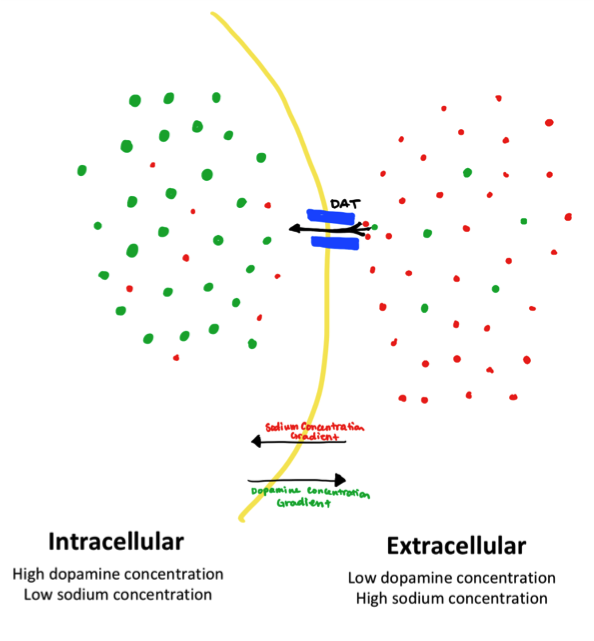
This concentration gradient is maintained by another membrane protein, the sodium potassium ion pump. This protein pumps three sodium molecules out of the cell as a step in ATP hydrolysis. This consistently maintains a sodium concentration of about 145 mmol /dm3 outside the cell and 5-15 mmol/dm3 inside the cell.9 This electrochemical gradient enables cotransport of dopamine with sodium via DAT.
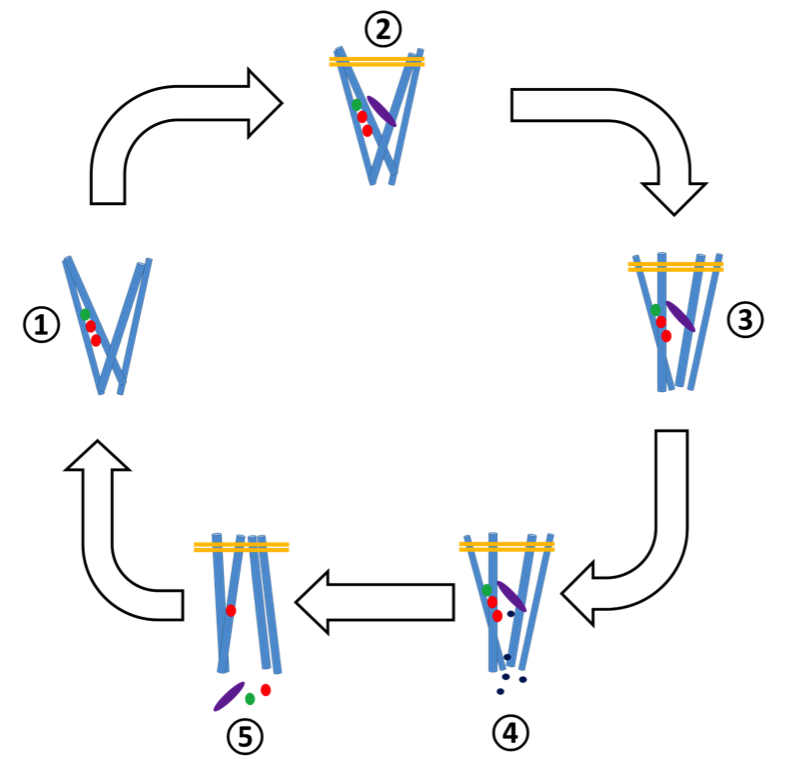
There are four binding sites in DAT that participate in transport: two sodium binding sites Na1 and Na2, one chlorine binding site Cl and one substrate binding site S1 . All are relatively close to one another in the molecule, located about halfway between the intracellular and extracellular regions.4
There are six steps of transport in DAT that proceed in the following order:4
- The transporter is outward facing (open to the extracellular region). Sodium and chloride ions diffuse into the molecule and bind in sites Na1, Na2 and Cl.
- Dopamine diffuses into the molecule and the hydrogen bond network is disrupted, allowing salt bridges to form between A85 and D476 and Y156 and F320. These bridges close the extracellular gate, keeping the dopamine inside the binding pocket. Simultaneously, two of the transmembrane regions begin to tilt inward.
- Because of these changes, the ligand cannot be accessed by either the intracellular or extracellular regions. This state is referred to as the holo-occluded state.
- Momentary dislocations of Na2 disrupt the hydrogen bonding network. This slightly opens the protein towards the intracellular region, allowing water molecules from inside the cell to leak into the binding sites. These waters destabilize the S1 site and cause the molecule to globally transition to the inward-facing conformation.
- In the reorientation, the Na1 and Cl binding pockets are enlarged, facilitating their release. Further, the protonation of D79 and D421 (by protons in the acidic intracellular environment) decrease the strength of their interactions with the dopamine in S1, releasing it to the intracellular environment. Na2 is not released.
- With no substrate bound, the extracellular gate reopens and the transmembrane regions tilt again to the outward facing conformation.
This process, which takes roughly 1 microsecond to complete, is visualized in figure 3 below.4
The role of chloride ions in this mechanism is not completely understood. It is clear that the ion is important in balancing charge, but may have other effects. One hypothesis is that when Cl- is bound, the affinity for Na+ in Na2 is lowered, which would allow for the momentary dislocations that lead to Na1, Cl, and dopamine release.5
Ion Binding Site Structure
There are two sodium ion metal centers in DAT. In both coordination sites, the charge of the sodium ion is 1+, the only biologically relevant oxidation state. Because sodium is not a transition metal. the ions have no d valence electrons (electron configuration 1s22s22p6), and thus primarily interact with the ligands through electrostatic interactions. Each ion is coordinated to five oxygen atoms in the protein chain. The ion in the Na1 binding site is also coordinated to one water molecule, creating an octahedral geometry. The Na2 site is not bound to a water, making it 5-coordinate and giving it a trigonal bipyramidal geometry. A water molecule is located 3.3 angstroms away from the sodium in Na2, in the secondary coordination environment.6
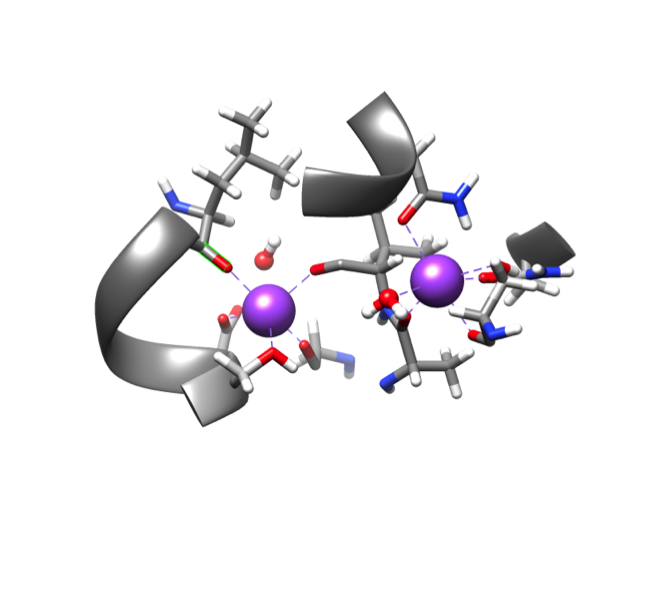
The three main factors that determine the structure of a coordination compound are the ligand field stabilization energy, sterics, and protein folding of the molecule. Because sodium ion has no d electrons, its Ligand Field Stabilization Energy (LFSE) is zero in any geometry, as demonstrated in figure 5.
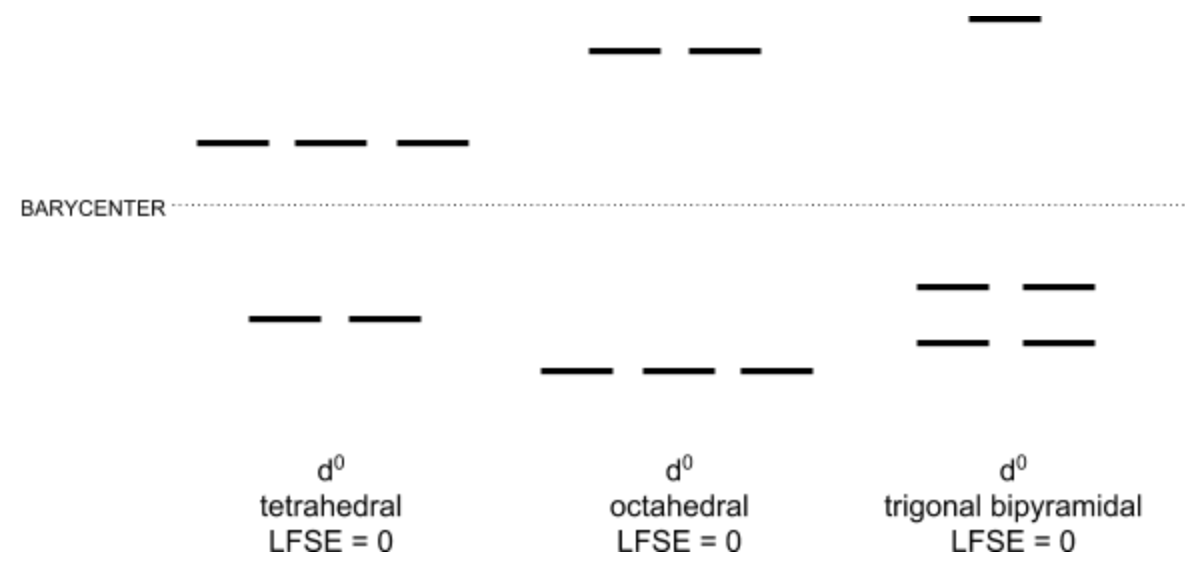
Because the LFSE is the same across all geometries, it doesn’t help in predicting the binding site geometry. The other factors that contribute to geometry are sterics and protein folding. The Na1 binding site is coordinated to 5 amino acid ligands and water, making it 6-coordinate, which is a relatively high coordination number, so sterics likely prevent further ligand binding. While sterics are relevant, protein folding is likely the driving force behind the geometry of the Na1 binding site. The structure of the protein as a whole dictates the binding site geometry. In the Na2 binding site, the metal ion is also bound to five amino acid ligands, but the water present is in the secondary coordination sphere. Again for Na2 as with Na1, protein folding is the dominating factor to select sodium binding site geometry because it has a zero LFSE and high coordination number.
Selection for Sodium
Sodium binding is integral to the function of DAT, so the protein must selectively bind sodium in a sodium and potassium-rich extracellular environment. Potassium and sodium are both alkali metals, so they have the same number of valence electrons and fairly similar properties.7 Because of their similar valence character, it is unlikely that this property is the criterion that the protein uses to selectively bind sodium. Furthermore, both ions most frequently coordinate with oxygen in amino acid residues.8 The preference for both potassium and sodium is explained by Hard-Soft Acid-Base theory. Both sodium and potassium are hard acids because they are not easily polarizable and tend towards ionic interactions. They prefer to interact with hard bases, like oxygen. Because both sodium and potassium are “compatible” with the same amino acids based on HSAB, ion hardness is also not a determining factor in the binding of sodium over potassium in DAT.
Neither valence electron character nor hard-soft acid-base character differ between sodium and potassium. However, the two ions do have different ionic sizes. Na+ has an ionic radius of 1.160 Angstroms, while K+ has an ionic radius of 1.520 Angstroms.10 This experimental evidence validates the theoretical prediction that the potassium ion is larger than the sodium ion because it is located below sodium in Group 1 in the periodic table. Because the potassium ion is larger than the sodium ion, it makes sense that the coordination lengths between a sodium ion and a residue would be shorter than the analogous potassium ion coordination lengths.
Ion size gives a clue into why the sodium ion will bind in this pocket and the potassium ion won’t. Perhaps the Na+ binding pocket in DAT is too small for the potassium ion. It is likely that the increase in the coordination length that would be required to coordinate the potassium would cause enough change in the molecule’s folding structure that it would no longer function properly. Further, this change in folding structure may make it impossible for the protein to make the correct conformational change to accomodate a K+ ion. If the energy required to make this conformational change (break disulfide bonds, alter hydrogen bonding structure) is greater than the energy gained by coordinating the metal ion, the coordination event will not occur. These predictions are consistent with experimental data in Marjorie Harding’s work, in which she found that sodium has lower overall coordination lengths with protein motifs than potassium does.8 Gouaux corroborated this hypothesis by noting that the mean coordination length of the sodium ion in the binding pocket is less than the mean coordination length of potassium with the same amino acids.6 The pairing of Harding and Gouaux’s work provides substantial evidence that ion size is a factor in the selective binding of Na+ over K+ in DAT.
Ion Lability
A labile coordination complex is one in which ligands are exchanged very quickly. In contrast, an inert compound is not labile and exchanges ligands slowly. The lability of a compound can be predicted using Taube’s rules by the strength of its ionic interactions and the d electron configuration. The stronger ionic interactions an ion has based on Coulomb's law (small, highly charged molecules have stronger interactions), the less labile it is. Na+, the ion in my system, has a small charge of 1+, making it fairly labile. This ion doesn’t have any d-electrons, which makes it very labile, according to Taube’s rules. Both evidence from the d-electron count and the Coulombic interactions indicate that Na+ is labile.
Also recall that the LFSE for Na(I) is zero. Zero is the largest possible LFSE value for an octahedral complex (all other values are negative). The more negative an ion’s LFSE, the more stable the complex is. Therefore, this large LFSE indicates that Na+ is thermodynamically unstable. Thermodynamic stability also informs chemical lability. It follows the trend that, generally, thermodynamically unstable compounds (based on LFSE) are labile. Applying this additional information to the sodium ion in the DAT system, we would predict that the Na+ is labile, just as Taube’s rules and Coulombic interactions would.
DAT can transport dopamine at a rate of roughly 1,000,000 molecules per second, so the quick release of sodium is good for the DAT system. If the metal weren’t labile, it would not release easily from the transporter, slowing down the overall transport process of dopamine.
Dopamine Transporter and Mental Health
DAT, and the related serotonin and norepinephrine transporters, are the targets of many drugs used to treat depression, ADHD, anxiety, Parkinson’s disease, and other neurological disorders. These drugs block reuptake of dopamine, that is, they inhibit the function of DAT, increasing the amount of dopamine in the synaptic cleft.2 It is also known that DAT is linked to drugs of abuse and addiction. For example, cocaine binds to DAT with high affinity, resulting in a high level of extracellular dopamine due to the inability of DAT to reuptake dopamine into the presynaptic neuron, which contributes to the euphoria cocaine users experience.1 Understanding how DAT works, particularly how it interacts with pharmaceuticals and recreational drugs, will lead to new approaches to treating prevalent psychological disorders, hopefully resulting in better patient health.1
References
- McHugh, P. C.; Buckley, D. A. Chapter Eleven - The Structure and Function of the Dopamine Transporter and Its Role in CNS Diseases. In Vitamins & Hormones; Litwack, G., Ed.; Hormones and Transport Systems; Academic Press, 2015; Vol. 98, pp 339–369
- Lin, Z.; Canales, J. J.; Björgvinsson, T.; Thomsen, M.; Qu, H.; Liu, Q.-R.; Torres, G. E.; Caine, S. B. Chapter 1 - Monoamine Transporters: Vulnerable and Vital Doorkeepers. In Progress in Molecular Biology and Translational Science; Rahman, S., Ed.; The Brain as a Drug Target; Academic Press, 2011; Vol. 98, pp 1–46.
- Nelson, N. The Family of Na+/Cl− Neurotransmitter Transporters. Journal of Neurochemistry 1998, 71 (5), 1785–1803.
- Cheng M. H.; Bahar, I. Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter. Structure 2015, 23 (11), 2171–2181.
- Borre, L.; Andreassen, T. F.; Shi, L.; Weinstein, H.; Gether, U. The Second Sodium Site in the Dopamine Transporter Controls Cation Permeation and Is Regulated by Chloride. J Biol Chem 2014, 289 (37), 25764–25773.
- Penmatsa, A.; Wang, K. H.; Gouaux, E. X-Ray Structure of Dopamine Transporter Elucidates Antidepressant Mechanism. Nature 2013, 503 (7474), 85.
- A. Reading The Periodic Table https://chem.libretexts.org/LibreTexts/Saint_Mary’s_College%2C_Notre_Dame%2C_IN/CHEM_342%3A_Bio-inorganic_Chemistry/Chapters/Chapter_1%3A_Atomic_Structure_and_the_Periodic_Table/1.2_The_Periodic_Table/A._Reading_The_Periodic_Table (accessed Jan 23, 2018).
- Harding, M. M. Metal–ligand Geometry Relevant to Proteins and in Proteins: Sodium and Potassium. Acta Cryst D, Acta Cryst Sect D, Acta Crystallogr D, Acta Crystallogr Sect D, Acta Crystallogr D Biol Crystallogr, Acta Crystallogr Sect D Biol Crystallogr 2002, 58 (5), 872–874.
- Crabb, E.; Moore, E. Metals and Life. RCS Publishing: Cambridge, England: 2010.
- Cordero, Beatriz, Veronica Gomez, Ana E. Platero-Prats, Marc Reves, Jorge Echeverria, Eduard Cremades, Flavia Barragan, and Santiago Alvarez. “Covalent Radii Revisited.” Dalton Transactions, no. 21 (2008): 2832–38.
Print this molecule!
Contributed by
This work was originally written by Leah Buck, Spring 2018: Leah is currently (as of 2018) a senior chemistry major at Saint Mary's College in Notre Dame, IN.
This work was originally edited by Dr. Kathryn Haas (Assistant Professor), Madison Sendzik (Teaching and Research Assistant), and Dr. Dorothy Feigl (Professor) at Saint Mary's College.


