Nickel Superoxide Dismutase (NiSOD)
- Page ID
- 97955
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Superoxide Dismutases (SODs) are enzymes that catalyze the dismutation, or removal, of superoxide. Superoxide is a dangerous oxygen radical that is often formed in aerobic pathways. Superoxide is similar to a dioxygen molecule. However, it contains an additional unpaired electron (Figure 1). This unpaired radical electron contributes a negative charge to the molecule. This radical is what makes superoxide dangerously reactive.1
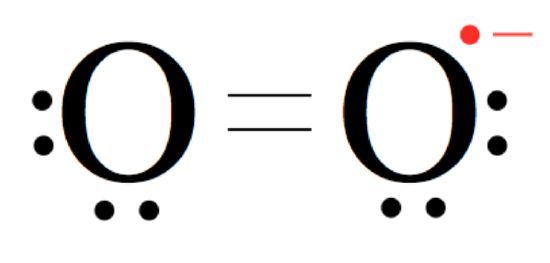
Figure 1. Structure of an oxygen radical, otherwise known as superoxide. Note the extra electron that constitutes the oxygen radical, contributing the overall negative charge, is depicted in red.
Superoxide is dangerous because it can react with DNA and disrupt the production of proteins that are essential for the life of the cell. Recent studies have even linked increased superoxide activity to devastating human diseases, including Alzheimer’s and Lou Gehrig’s Disease (ALS). The dismutation of oxygen is essential in protecting the cell against oxidative damage and preventing such devastating diseases, promoting functional cellular processes. SODs are important because they are an organism’s first defense against harmful superoxide anions. Specifically, SODs catalyze the superoxide dismutation, which consists of the simultaneous oxidation and reduction of oxygen radicals, or superoxide, to produce hydrogen peroxide (H2O2) and molecular oxygen (O2) at rates near the diffusion limit of the cell. The dismutation reaction of superoxide, catalyzed by SOD, is shown in Figure 2. This reaction converts the dangerous superoxide radicals to less reactive products.
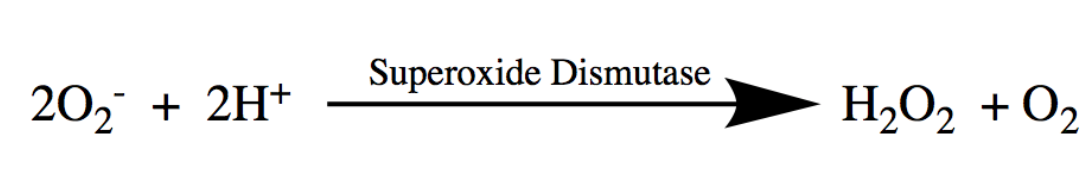
Figure 2. Superoxide Dismutase catalyzes the dismutation of superoxide. In the dismutation reaction, the oxygen radical, superoxide, reacts with hydrogen ions (H+), catalyzed by superoxide dismutase, to form hydrogen peroxide (H2O2) and dioxygen (O2), converting dangerous superoxide radicals to less reactive products.
There are multiple classes of SOD enzymes. Nickel superoxide dismutase (Ni-SOD), portrayed in figure 3, is a relatively recently discovered class of SOD. Compared to other members of the SOD family, Ni-SOD is unique. For instance, Ni-SOD is not found in humans. Though it is present in some eukaryotic cells, such as various algae species, Ni-SOD is most often found in bacteria. Ni-SOD molecules are primarily present in the cytoplasm of the cell.1,
The structure of Ni-SOD is important in catalyzing superoxide dismutation. Ni-SOD is a product of the sodN gene and is comprised of a 117 amino acid sequence that has no homology to other SODs. Ni-SOD is a globular protein. In other words, the overall shape of the protein is relatively spherical. As is common with globular proteins, Ni-SOD is soluble in aqueous solutions. In addition to its spherical shape, it has a hollow center and is considered a homohexamer, that is, it is comprised of six identical subunits.1,3 This structure is depicted in figure 3 below.
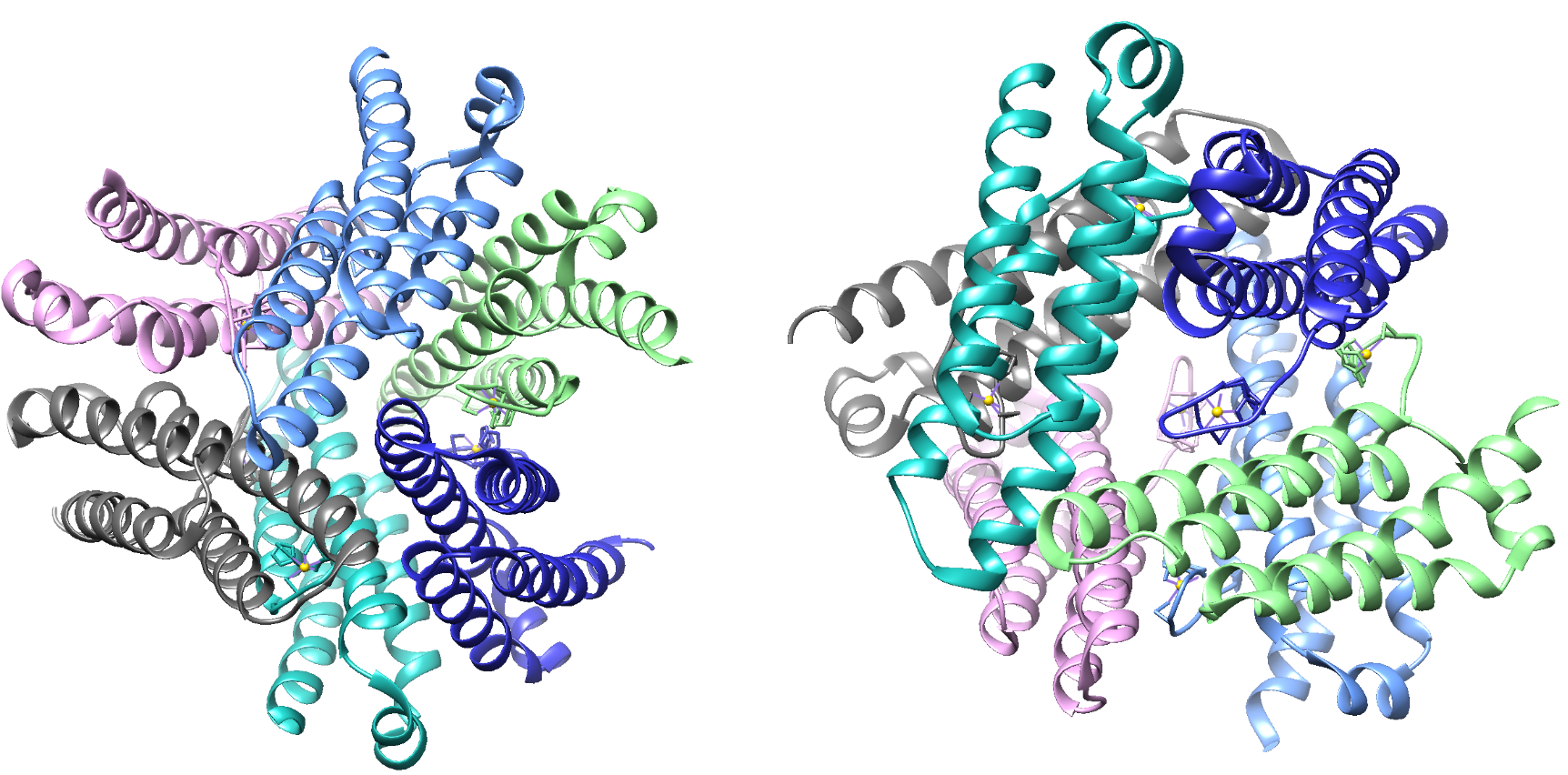
Figure 3. Two different perspectives of the globular (spherical) molecular structure of Ni-SOD (PDB: 1T6I). The structure on the left emphasizes the hexamer structure of Ni-SOD. Each of the six identical subunits is portrayed in a different color. The structure on the right portrays the hollow center of Ni-SOD.
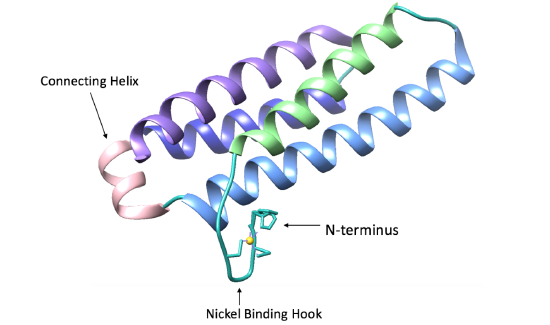
As seen in figure 4 below, each Ni-SOD subunit has a right-handed fold consisting of four alpha helices, following an up-down-up-down pattern. In addition, there is also a short connecting helix (shown in pink, figure 4). This connecting helix is common for 4-helix bundles and aids in structural stability of the bundle.1
|
|
As its name suggests, Ni-SOD’s catalytic activity is dependent upon a nickel cofactor in its active site. The nickel binding motif, from crystal structure data, is shown in figure 5 below. This histidine appears in two conformational changes that depend on the oxidation state of nickel. The nickel cofactor participates in the Ni-SOD system in its Ni2+ and Ni3+ oxidation states. Nickel binds to Ni-SOD at the N-terminus, at a site referred to as the “nickel binding hook”, shown in figures 4 and 5. This name comes from the curved structure of the nickel binding site on each monomer of Ni-SOD, which resembles a hook. The nickel binding hook is located within the first 6 residues from the N-terminus of each of the subunits (figures 4 and 5). Studies have proposed that the nickel binding motif is N-terminal His- Cys- X- X- Pro- Cys- Gly- X- Tyr. This nickel binding hook contains all of the nickel binding amino acid residues, or ligands, as well as the superoxide substrate stabilizing ligands. Studies have shown that the nickel binding hook motif accounts for all of Ni-SOD’s activity. 6 This nickel binding hook makes up the active site of Ni-SOD, where superoxide is dismutated. This active site is unique, comparatively, because many other human SODs contain pre-formed active sites. That is, their active sites, in which superoxide binds, are not dependent on the binding of the metal cofactor. Each of the six subunits, or monomers, of Ni-SOD contains an active site, or nickel binding hook.1. |
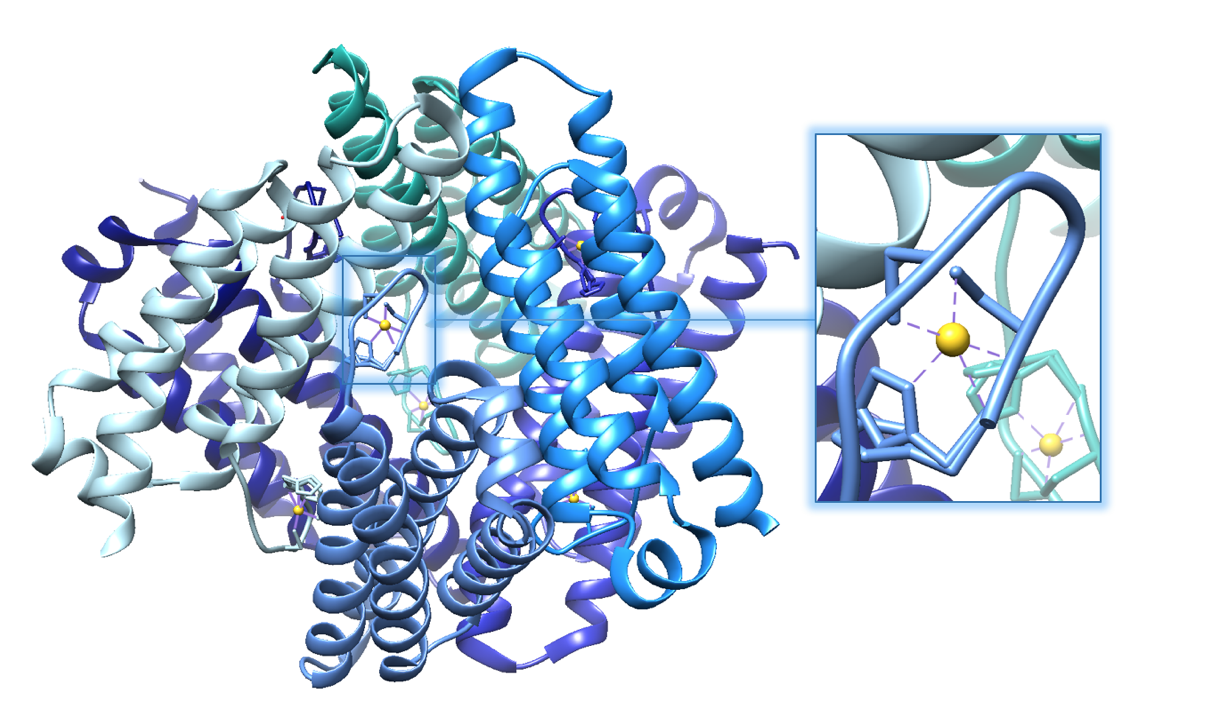
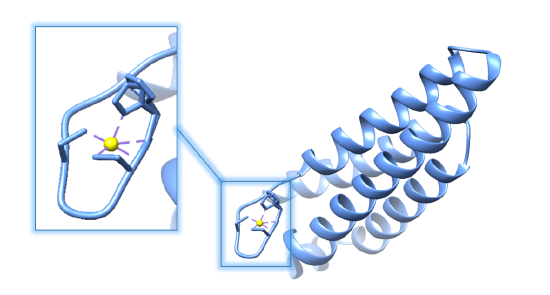
A. B.
Figure 5.A. Structure of Ni-SOD (PDB: 1T6I). The active site (Ni-SOD complexed with the nickel cofactor) is emphasized in the figure. Nickel cofactors are shown in yellow. The structure of Ni-SOD contains 6 subunits, or monomers (shown in different shades of blue), each containing a nickel cofactor. There are 6 nickel cofactors bound, one Ni active site in each of the six monomers. B. The monomer of Ni-SOD with the bound nickel cofactor emphasized.
The dismutation of toxic superoxide molecules via Ni-SOD is dependent upon the binding of a nickel cofactor. Therefore, the binding of this nickel cofactor, which forms the structure of the Ni-SOD active site, is essential to Ni-SOD’s function. As was stated, the nickel binding hook motif (N-terminal His- Cys- X- X- Pro- Cys- Gly- X- Tyr) contains all of Ni-SOD’s nickel binding ligands, including the N-terminal amine and imidazole nitrogen of His-1, the N-terminal amine of Cys-2, and the side chain sulfurs of Cys-2, and Cys-6.,, These nickel binding ligands, particularly Cys-2 and Cys-6, are uncommon donor groups for superoxide dismutase proteins in general. This is due to the fact that Cysteine residues are prone to oxidation, due to their tendency to form disulfide bonds and sulfoxides, leading to a high risk of oxidative damage. Ni-SOD is the only SOD that has been studied that contains sulfur-donor ligands, the cysteines, in the active site. The importance of these uncommon cysteine ligands will be discussed below. Though these cysteine residues are uncommon in their binding to Ni, they are essential to Ni-SOD’s function.2,6
Many factors account for the high binding affinity of Ni-SOD for nickel ions, one being the chelate effect. In molecular systems, ligands can be classified as monodentate, bidentate, or polydentate. Monodentate ligands only have one atom capable of binding to a central metal ion or atom, while bidentate ligands have two atoms capable of binding, and polydentate ligands have more than two atoms capable of binding. Bidentate and polydentate ligands are referred to as chelating ligands, while monodentate ligands are non-chelating ligands The term “chelate” comes from the concept of a “claw.” A chelator is defined as a ligand that can be bound to a central atom at two or more points, effectively “grabbing” the central atom like a “claw”. The chelate effect refers to the increased affinity of chelating ligands for a specific metal ion versus that of non-chelating ligands. This effect plays a role in the Ni-SOD system. Nickel initially binds to the nickel binding hook in its Ni2+ oxidation state at four binding sites, including the N-terminal amine of His-1, the N-terminal amine of Cys-2, and the side chain sulfurs of Cys-2, and Cys-6, which is depicted in figure 6 below. Therefore, each hook is considered a multidentate chelator. This contributes to the Ni-SOD’s high binding affinity for nickel because the chelate effect states that there is an increased affinity of chelating ligands for a specific metal ion versus that of non-chelating, or monodentate, ligands. Chelation is an entropic benefit that increases stability. This increase in entropy effectively increases the hook’s affinity for nickel, allowing the protein to sequester nickel from other nickel binding molecules in the cytoplasm.6,7
|
In addition to the chelate effect, Hard Soft Acid Base (HSAB) theory also plays a role in the binding affinity of nickel metal ions to Ni-SOD. The HSAB character of certain ligands in a system can help select for the correct metal ion. HSAB theory specifies the “hard’ or “soft” character of Lewis acids and bases. The hard or soft character is based on charge density, polarizability, and the types of interactions that are occurring. For example, atoms with a more “hard” character tend to participate in more electrostatic interactions, while atoms with more “soft” character tend to participate in more covalent interactions. Since metals ions, which are Lewis acids and ligands are Lewis bases, HSAB theory can be applied to coordination compounds. More specifically, HSAB theory states that larger, more soft, in other words, more polarizable, ligands tend to have a higher affinity for larger, more soft metals. Conversely, smaller, more hard, in other words, less polarizable, ligands have a higher affinity for smaller, more hard metals. In the simplest of terms, “like prefers like”.7 The choice of ligands in the active site of Ni-SOD is different than those found in other superoxide dismutases. However, the choice of ligands can be partly explained by Hard-soft acid-base theory and the fact that the nickel cofactor is involved in the system as multiple oxidation states, cycling between Ni2+, which has borderline character and Ni3+, which has hard character. In such an active site, a mixture of hard and borderline donor atoms may be expected, and in fact this is what is found with the hard nitrogen donor of the N-terminus and the borderline His-1 imidazole side chain. The superoxide ligand that binds here is a hard donor and is, therefore, a good match for the Ni3+ oxidation state. However, there is the unexpected appearance of the soft thiolate donors from Cys-2, and Cys-6. Although the soft donors of Cys are uncommon in SOD’s and are unexpected based on hard-soft acid-base theory, they are essential to tuning Ni-SOD’s redox potential.2,6, This will be discussed later in more detail. |
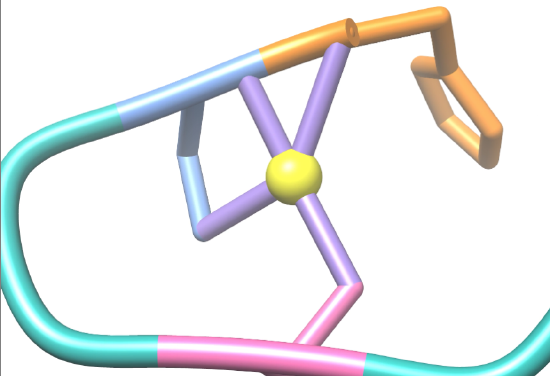
Figure 6. Ni-SOD in its complexed form with the Ni2+ cofactor (PDB: 1T6I). Each nickel binding hook acts as a multidentate chelator for Ni. The Ni2+ metal center (yellow) and its binding sites, His-1 (orange), Cys-2 (blue), and Cys-6 (pink) are shown here. Notice the multiple donor groups, or ligand bonds, in the coordination environment of nickel in the active site, represented by the purple nickel-ligand bonds. This is indicative of a multidentate chelator.6,7 |
The binding affinity of Ni-SOD for nickel can also be described by Ligand Field Theory (LFT). The abbreviated electron configuration of a nickel metal atom is [Ar]3d84s2. The electron configurations of Ni2+ and Ni3+ are [Ar]3d84s0 and [Ar]3d74s0 , respectively. In the ions, the valence electrons occupy the 3d-orbital, and not the 4s orbital because the 4s orbital is a higher energy orbital. Ni2+ and Ni3+ are 3d8 and 3d7 metals, respectively. That is, Ni2+ and Ni3+ have 8 and 7 d-electrons, respectively.7
According to LFT, d-orbital electrons correspond to either high or low energy levels. Higher energy d-electrons correspond to anti-bonding electrons, while lower energy d-electrons correspond to non-binding electrons. LFT d-electron splitting can be used to describe the high or low spin of electrons. Δ signifies the distance between the high and low energy levels. Higher Δ values correspond to strong fields, while lower Δ values correspond to weak fields. It requires less energy, and is, therefore, easier, for d-electrons to “jump” from the lower energy level to the higher energy level in weak fields because the distance between the two levels is smaller. Therefore, d-electrons in weak fields with low Δ values tend to jump to the higher energy level before pairing up because it requires less energy, while d-electrons in strong fields with high Δo values tend to pair up first because it requires less energy than jumping to the higher energy level. Metal complexes with strong fields are referred to as low spin, and metal complexes with weak fields are referred to as high spin.7
As can be observed by the ligand bonds in figure 6 above (shown in purple), Ni2+ binds to Ni-SOD in a square planar geometry. This is expected for a d-8 metal based on ligand field theory. This Ni2+ metal center is low spin, due the fact that square planar geometries have relatively large delta (Δ) compared to other geometries, as seen by the comparison of square planar and tetrahedral d-electron splitting diagrams in figure 7 below. As was previously discussed, considering the large Δ of Ni2+’s square plane, this corresponds to a strong field and low spin. Typically, strong field ligands are associated with low spin metal centers and weak field ligands are associated with high spin metal centers, however, since square planes are generally low spin due to their large Δ values, the weak or strong field nature of the nickel binding ligands of Ni-SOD do not play a major role in the determining the spin of the Ni2+ metal center.6,7
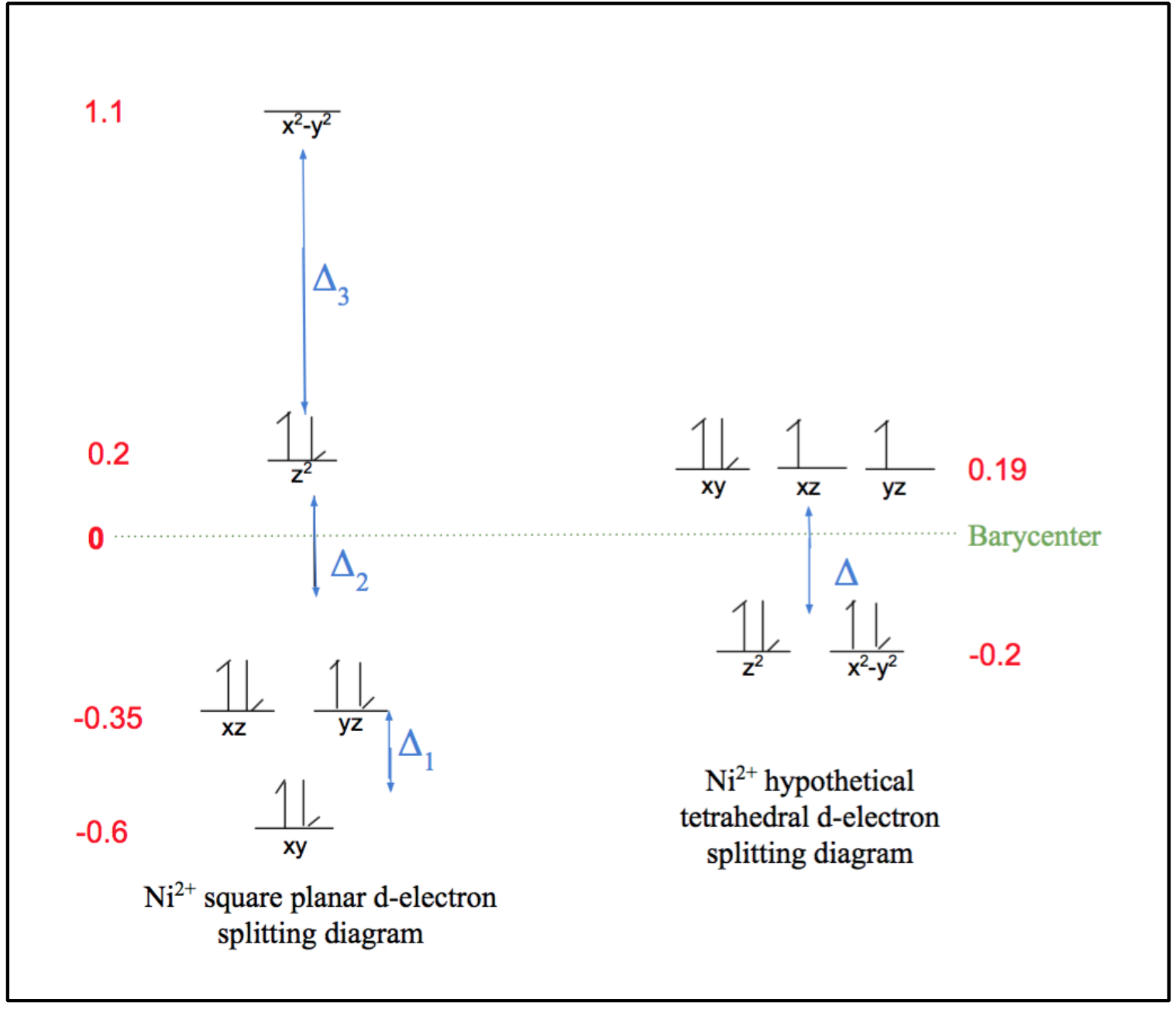
Figure 7. From left to right, d-electron splitting diagrams of the Ni2+ square planar geometry and the hypothetical Ni2+ tetrahedral geometry. The green dotted line corresponds to the barycenter, which is associated with an energy value of zero, red numbers correspond to the energy values of each d-electron energy level, and the blue deltas indicate the energy differences between the lower and higher energy levels. Notice that the square planar geometry has significantly greater, or more spread out splitting, corresponding to a greater Δ, than that of the tetrahedral geometry.
Also due to its large splitting energies, the square planar geometry maximizes ligand field stabilization energy (LFSE) for d-8 metals. This energy benefit causes the square planar geometry to be favored because LFT states that the metal complex will employ the configuration with the greatest, most negative, LFSE, which corresponds to the greatest stability.7 This energy benefit of a square plane relative to a tetrahedron is shown by the calculations highlighted in equations 1 and 2. As seen in these equations, LFSE is calculated by determining the sum of the energy differences between the high and low energy levels (Δ). Note that LFSE is a negative value, thus the greatest LFSE corresponds to the most negative value.
Square Plane: LFSE = ((-0.62*2) + (-0.35*4) + (0.2*2) +1.1)Δ = -2.2Δ (1)
Tetrahedron: LFSE = ((-0.2*4) + (0.19*4))Δ = -0.04Δ (2)
Ni-SOD is a redox catalyst. Therefore, its mechanism of superoxide dismutation is driven by redox reactions in which the redox active Ni cycles between oxidized and reduced states (Ni3+ and Ni2+, respectively), differing by one electron. This is common to SODs.2,6 These reactions are shown below. Equation 3 portrays the simultaneous reduction of nickel and oxidation of superoxide (O2-), equation 4 represents the simultaneous oxidation of nickel and reduction of superoxide, and equation 5 portrays the overall dismutation reaction of superoxide.
Ni3+ + O2- → Ni2+ + O2 (3)
Ni2+ + O2- + 2H+ → Ni3+ + H2 O2 (4)
2O2- + 2H+→ O2 + H2 O2 (5)
These reactions are dependent upon a redox active nickel cofactor. However, nickel does not catalyze superoxide dismutation in neutral, aqueous solutions. For instance, other common SOD cofactors are more redox active and can cycle between their oxidation states in biological fluids, thus dismutating superoxide, while Ni does not dismutate superoxide in the absence of the Ni-SOD protein. This is due to the fact that the reduction potential of Ni3+ to Ni2+ is proposed to be ~2V near pH~7.2 This value is much larger than the proposed acceptable range for catalysis of superoxide dismuatation, which includes reduction potentials ranging from -0.160-0.870V near pH~7.2
In order to account for this large reduction potential, Ni-SOD utilizes uncommon active site ligands, Cys-2 and Cys-6, to tune the reduction potential of Ni3+ to Ni2+ to ~0.30V (pH~7) , which falls in the acceptable range for catalysis (-0.160-0.870V near pH~7). It is proposed that these Cys residues tune Ni reduction potential through bonding. The covalent nature of the Ni-S(Cys) bonds is due to the ��-bonding interactions. In addition, these ligands cause Ni2+-based oxidation to be favored over S-based oxidation and assist in tuning the reduction potential of Ni. Moreover, studies have proposed that the S-donor of Cys-2 is the protonation site, or the proton (H+) source, for the oxidative half of the reaction, due to the greater electron density of its side chain, compared to that of Cys-6.2 Therefore, due to redox tuning via its active site ligands, nickel is a redox active metal ion when it is bound to Ni-SOD, making superoxide diamutation possible.2,6
Once Ni2+ is bound to Ni-SOD, tuning the reduction potential of nickel and forming the active site, superoxide dismutation can occur. Ni-SOD facilitated superoxide dismutation occurs via the catalytic, cyclic mechanism (figure 8) that is driven by the aforementioned redox reactions. The steps of this cycle can be defined as electron transfer steps, indicating the presence of redox reactions, and/or ligand association of dissociation steps, indicating the binding or dissociation, respectively, of the superoxide substrate. Electron transfer in Ni-SOD’s catalytic cycle occurs between the superoxide substrate and the active site Ni, to which it binds. Inner sphere mechanisms refer to mechanisms in which the metal is oxidized or reduced by a ligand bound directly to the metal, while outer sphere mechanisms employ redox reactions more indirectly, that is, via ligands that are not directly bound to the metal center. Therefore, Ni-SOD’s mechanism is considered an inner sphere mechanism.6,7
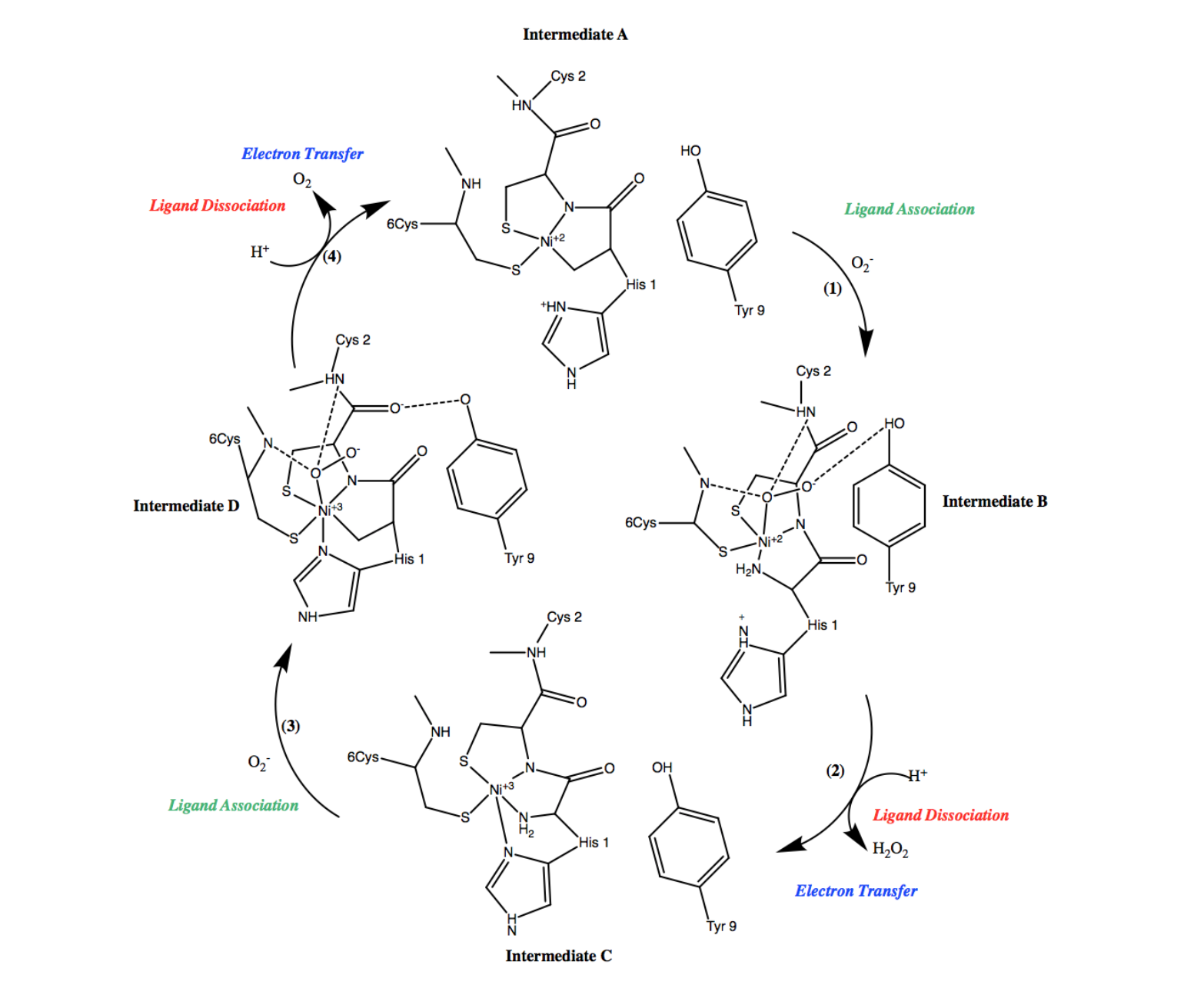
Figure 8. Cyclic mechanism of Ni-SOD facilitated dismutation of superoxide. Note that the cycle contains 4 steps. Ligand association steps are indicated in green, ligand dissociation steps are indicated in red, and electron transfer steps are indicated in blue. In addition, each of the 4 intermediates of the cycle are also labeled A-D.
As shown in the first step of the Ni-SOD catalytic cycle (figure 8, (1)), an oxygen radical, or superoxide, is introduced and binds to Ni2+ in the active site. In the second step of the cycle (figure 8, (2)), the oxygen radical is then reduced to hydrogen peroxide (H2O2), thus dismutating the superoxide and the nickel ion is simultaneously oxidized from Ni2+ to Ni3+. As was previously discussed, the proposed source of the protons in this step is the Cys-2 residue.2
During this step, in the oxidation from Ni2+ to Ni3+, the metal binding hook undergoes a conformational change (figure 9), in which His-1 is coordinated to bind to Ni3+, resulting in an additional ligand bond between Ni3+ and the imidazole N of His-1.4,6
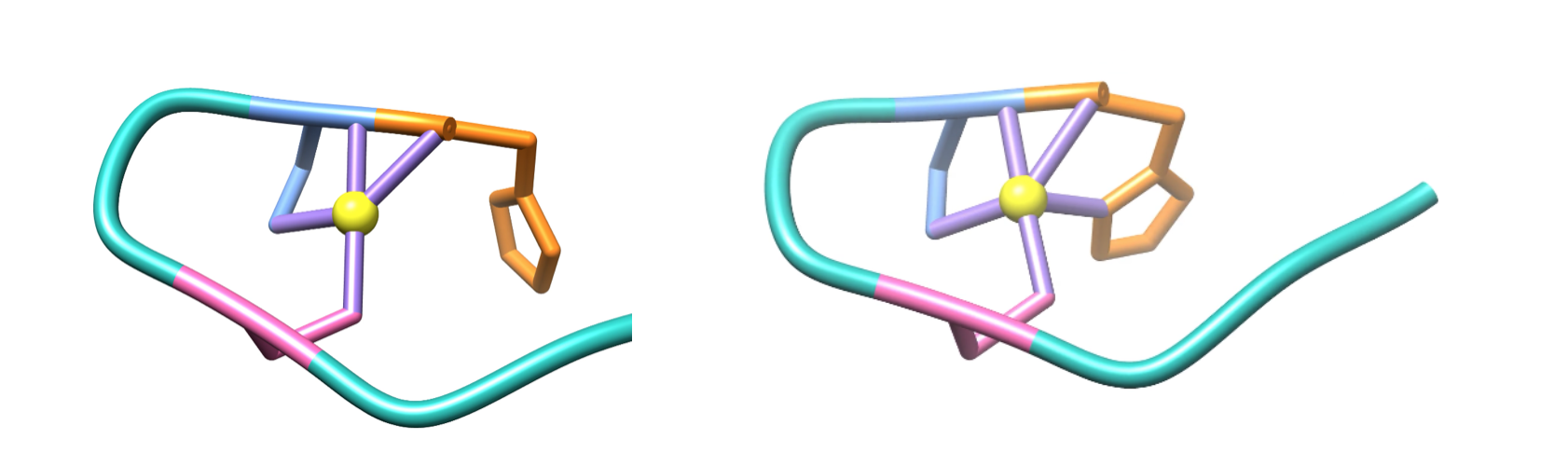
Figure 9. From left to right, the coordination environments of the nickel cofactor are shown before and after the conformational change that occurs during the second step of the Ni-SOD facilitated dismutation of superoxide (PDB: 1T6I). The nickel cofactor (yellow) is in its Ni2+ oxidation state on the left and is oxidized to its Ni3+ oxidation state on the right. His-1 is shown in orange, Cys-2 is shown in blue, and Cys-6 is shown in pink. Note that, as seen by the coordination environment on the right, this conformational change coordinates the imidazole N of His-1 to bind to Ni3+, forming an additional ligand bond, therefore, changing the geometry from square plane (left) to square pyramid (right).
This conformational change causes the geometry of the Ni3+ coordination environment to change from the square plane of the Ni2+ coordination environment to a square pyramid, as shown in figure 9 above. This geometry is also expected based on LFSE. Similar to Ni2+’s square plane, this square pyramid geometry of the Ni3+ coordination environment has similar large splitting energies, or Δ, corresponding to a strong field and low spin, as seen in the d-electron splitting diagrams in figure 10 below.7
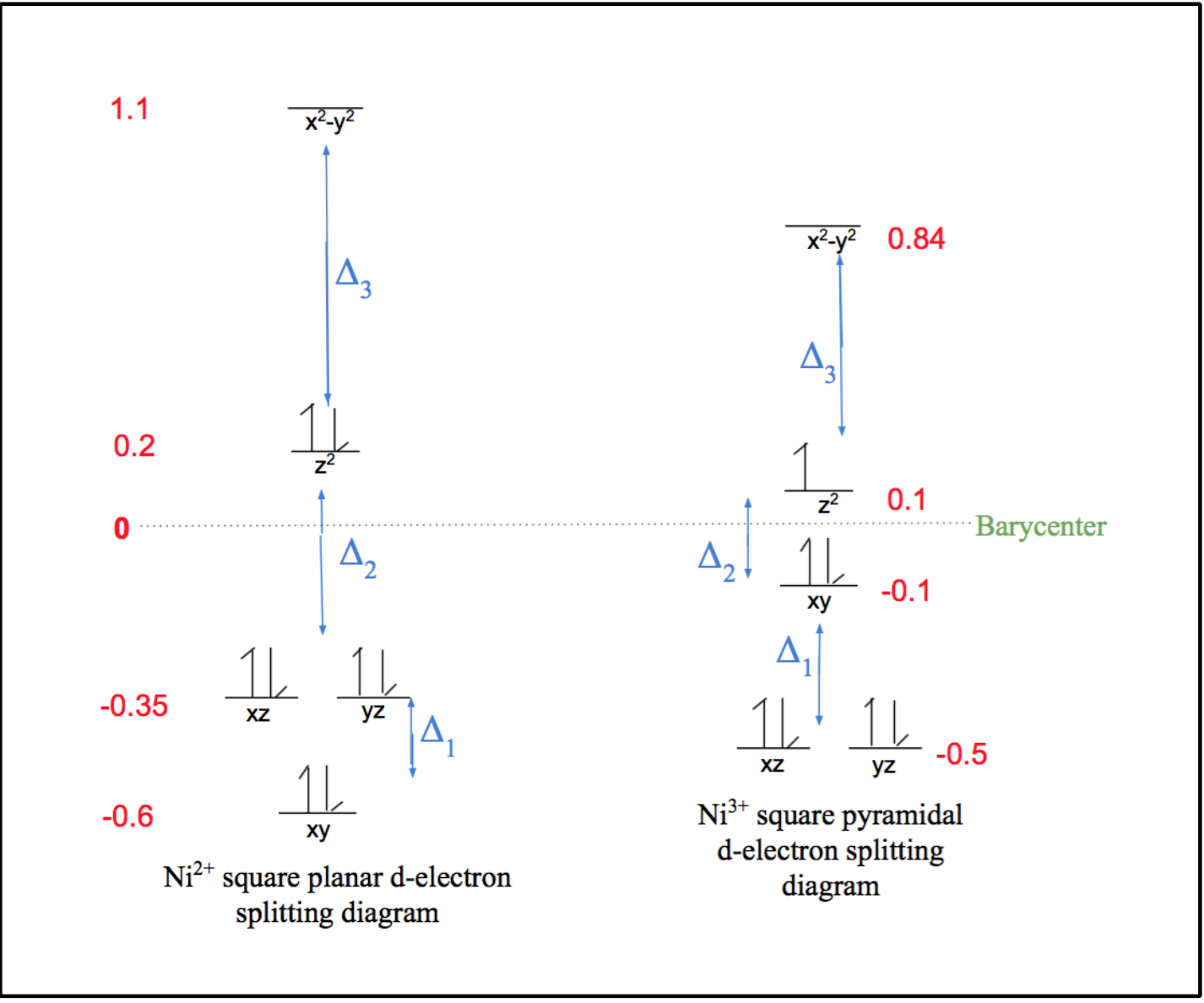
Figure 10. From left to right, d-electron splitting diagrams of the Ni2+ square planar geometry and Ni3+ tetrahedral geometry. The green dotted line corresponds to the barycenter, which is associated with an energy value of zero, red numbers correspond to the energy values of each d-electron energy level, and the blue deltas indicate the energy differences between the lower and higher energy levels. Notice that the square pyramid geometry has similar, spread out splitting, corresponding similar to large Δ values.
Therefore, similar to the square plane of Ni2+, the square pyramid geometry of Ni3+ is expected due to its increased LFSE benefit, which corresponds to increased stability, as shown by the calculations shown in equations 6 and 7 below.
Square Pyramid: LFSE = ((-0.5*4) + (-0.1*2) + (0.1*1) +0.84)Δ = -2.0Δ (6)
Square Plane: LFSE = ((-0.62*2) + (-0.35*4) + (0.2*2) +1.1)Δ = -2.2Δ (7)
During the third step of Ni-SOD’s catalytic cycle (figure 8, (3)), another superoxide molecule is introduced to the system and binds at the Ni3+ in the active site. In the fourth, and final step of the cycle (figure 8, (4)), the superoxide is dismutated via oxidation to dioxygen (O2). Similar to step 2, the proposed source of the protons in this step is the Cys-2 residue.2 Simultaneously, the nickel ion is reduced from Ni3+ to Ni2+, during which an opposite conformational change occurs, resulting in the imidazole of His-1 no longer being coordinated. At this point, the active site has returned to the form of intermediate A and the cycle continues as Ni-SOD functions to dismutate superoxide. Each step of its catalytic cycle is essential for the function of Ni-SOD.6
Ni-SOD’s catalytic cycle can be even further understood by considering the 18-electron rule, which states that the metal centers are most stable with electron counts of 18, followed by an electron count of 16. Electron count of metal complexes refers to the number of valence electrons present on the central atom, and it includes both the metal d-electrons and the electrons that are part of coordinate covalent bonds. The total electron count is calculated by adding the number of electrons shared by ligand bonds to the central metal ion (2 electrons per bond) to the number of metal d electrons. The number of d electrons are calculated from the oxidation state of the central metal ion. Recall that Ni2+ and Ni3+ are d8 and d9 metals, respectively. For instance, Ni2+ has a formal charge of +2, and thus it has 8 d-electrons. Taking this into consideration, along with the electrons that are part of coordinate covalent bonds, the electron count of intermediate A of Ni-SOD’s catalytic cycle is 16 e-, intermediate B is has an electron count of 18e-, intermediate C has an electron count of 17e-, and intermediate D has an electron count of 19e-. As expected, species that are 18 electrons or above, proceed to dissociation of a ligand, while species that are less than 18 electrons gain electrons in a ligand association step, driving the cyclic superoxide mechanism forward (figure 11).5,7
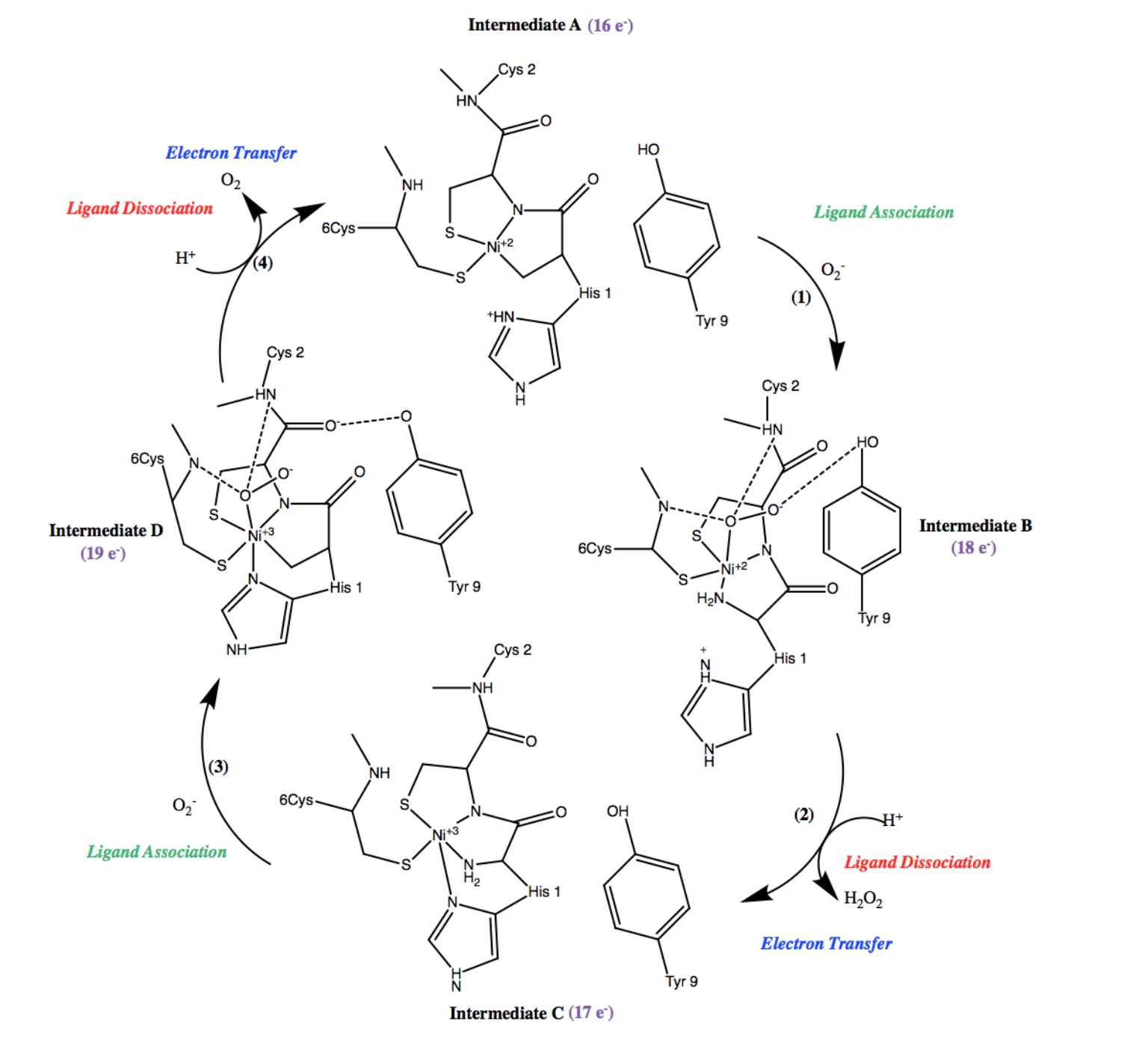
Figure 11. Cyclic mechanism of Ni-SOD facilitated superoxide dismutation. Notice that the electron counts of each intermediate are labeled in purple. Intermediate A has 16 e-, intermediate B is has an electron count of 18e-, intermediate C has an electron count of 17e-, and intermediate D has 19e-. As expected, species that are 18 electrons or above, proceed to dissociation of a ligand, while species that are less than 18 electrons gain electrons in a ligand association step, driving the cyclic superoxide mechanism forward.5,7
|
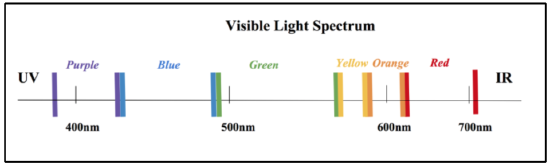
Figure 12. The visible light spectrum. Note that the visible light spectrum spans from ~ 400 - 700 nm, moving from purple to red as distances increase (nm). The UV region is below the visible light spectrum and the IR region is above it. Each of the regions corresponding to specific colored light absorbances are labeled here. |
In order to further observe Ni-SOD, analysis methods are necessary. One effective method of analyzing Ni-SOD is UV-Vis Spectroscopy. Certain geometries, or metal complexes, correspond to specific colors. These metal complexes can be analyzed by UV-vis spectroscopy because they absorb in the visible light region, which includes wavelengths spanning from approximately 400 to 700nm, as is depicted by the visible light spectrum in figure 12 below. In recent studies, Ni-SOD has produced a strong absorbance at around 260 nm. This absorbance corresponds to an extinction coefficient of �� ≈ 10,000 to 13,000 M-1cm-1. When studying metal complexes, two different types of electronic transitions that can be observed with UV-vis light: d-d transitions and charge transfer transitions. d-d transitions can only occur when the metal has partially filled d-orbitals (that is d1-d9 metals). These transitions are excitations of electrons in the d orbitals, from low to high electron energy levels, crossing Δ. These are less intense, that is, they correspond to a smaller extinction coefficient (�� less than 1000 M-1cm-1). Charge transfer transitions are transitions that occur independent of d-electrons. These are more intense and correspond to larger extinction coefficients (�� much greater than 1000 M-1cm-1). |
Charge transfer transitions can occur in two ways: from metal to ligand and from ligand to metal. Metal to ligand transitions often include high charge metals and require ��-acceptor ligands, while ligand to metal is more common and requires ��-donor ligands.6 Since the nickel oxidation states involved in the Ni-SOD system, Ni2+ and Ni3+, are d8 and d7 metals, respectively, these could, potentially, correspond to d-d absorptions transitions. However, considering the fact that Ni-SOD’s absorption at 260 nm is so intense, corresponding to a high extinction coefficient (�� ≈ 10000 to 13000 M-1cm-1), and that this absorbance corresponds to the protein as a whole, rather than just the nickel cofactor, this indicates that this is a charge transfer absorption.7.
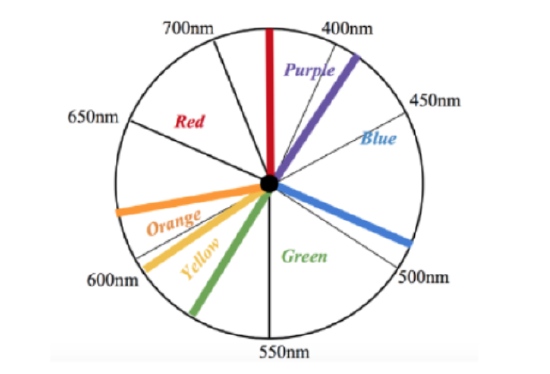
As stated the absorption at 260 nm is not in the visible light spectrum, however, this absorbance corresponds to the Ni-SOD molecule as a whole, so it is not indicative of the nickel metal complex. In addition to this absorption, studies have also found a very slight absorbance via Ni-SOD around 460 nm, which has been proposed to be related to the nickel complex. Though these studies have not found the extinction coefficient of this absorption, its weak nature indicates that this absorption is a d-d transition. Though this is a weak absorbance, it is useful because this d-d transition could be used to measure Δ and if it is measurable, it is useful. Since this absorbance is within the visible light spectrum, it can be analyzed using Uv-vis spectroscopy. As can be observed from the visible light spectrum above (figure 12), this 460nm absorbance corresponds to blue light. This weak absorbance also indicates that the nickel metal center would be slightly colored. As can be determined by the color wheel shown in figure 13 below, metal centers display colors that oppose the the color light that they absorb. Therefore, it would be expected that, if concentrated, the Ni complex of Ni-SOD would have a light yellow-red color, tending more toward yellow.7,9,10
|
Figure 13. Visible light spectrum color wheel. Each color region is labeled. Note that metal centers display colors that oppose the the color light that they absorb.7. |
In summation, Ni-SOD molecules play an essential role in the health of the cell through their antioxidant activity. The function of these powerful molecules and their dependence on nickel cofactors has been described here. Further research pertaining to Ni-SOD and its function is important because it will further understanding of the essential role of this unique superoxide dismutase and its significant effects on cellular processes.