Introduction to Bioconjugation
- Page ID
- 59317
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Bioconjugation is a chemical technique used to couple two molecules together, at least one of which is a biomolecule, such as a carbohydrate, nucleic acid, or protein. Proteins are especially diverse biomolecules due to the variety of amino acids available and are thus important substrates in bioconjugation reactions. Bioconjugation reactions play a critical role in the modification of proteins. Because of recent advances in the study of biomolecules, proteins can be modified to perform a variety of functions, including cellular tracking, imaging biomarkers, and target drug delivery.
Synthesis and Strategies
Though the chemistry behind conjugation is simple, its execution is not. There are several obstacles that can hinder efficient bioconjugation reactions. For example, because certain amino acid residues are more prevalent than others, some reactions cannot be selective and are therefore inefficient. Another obstacle is polar molecules on the surface of proteins that can interfere with a reaction. In addition, selectively modifying more than one site on a protein presents considerable challenges.
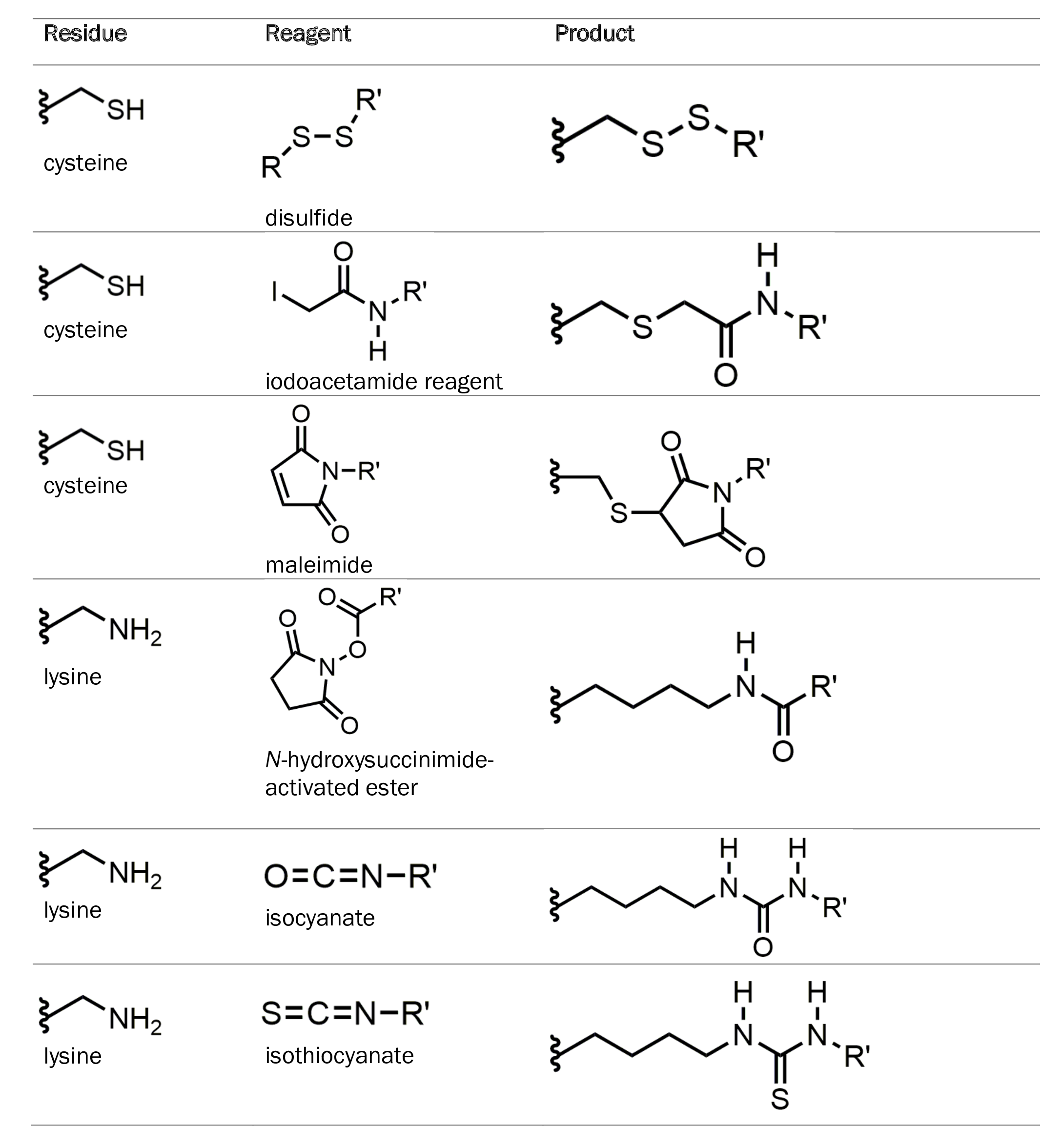
Classical approaches to protein modification are typically second-order reactions that target side chains of specific amino acids such as cysteine and lysine. Cysteine and lysine side chains contain thiol and amino groups, respectively, which allow them to undergo modification with a wide variety of reagents. The table below summarizes common reactions involving these amino acids.
However, with the advent of new technologies and advancements in biochemistry, strategies have been developed to make bioconjugation reactions more efficient.
Choosing a strategy is largely dependent on the protein of interest. If the protein is present in a mixture and cannot be isolated, alternative techniques will have to be used. If the protein is present in its purified form, the next question to consider is whether site specificity is needed. From these initial criteria, a flowchart, developed by Nicholas Stephanopolous and Matthew Francis, can be used to determine efficient synthesis techniques on a case-by-case basis.
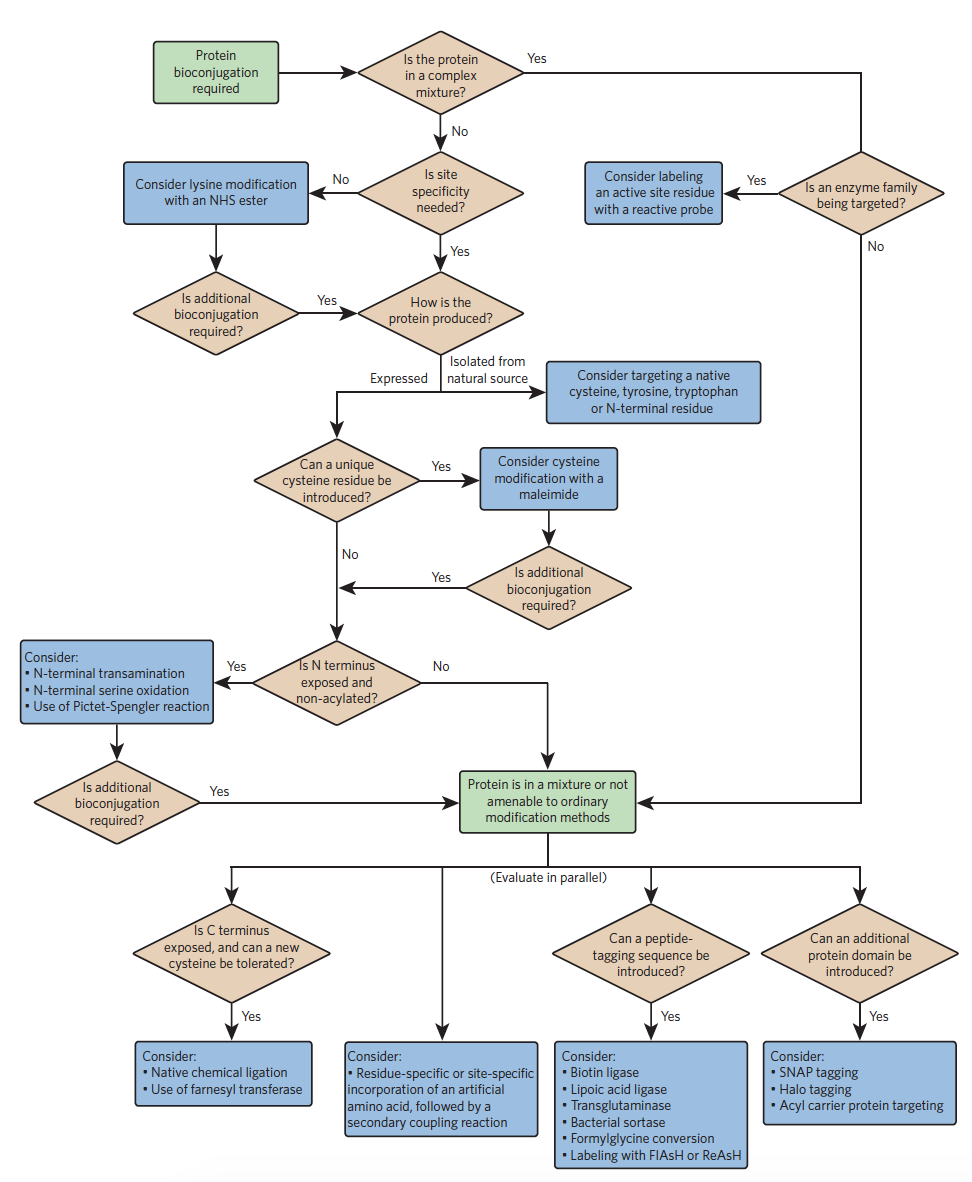
Some common bioconjugation techniques introduced in this flowchart are discussed below.
Bioorthogonal Reactions
One subset of bioconjugation reactions which has become increasingly important is bioorthogonal reactions, which are reactions in living systems that do not interfere with native processes. They provide a mechanism for targeting and modifying a specific site on a protein. Applications include live cell surface labeling, enhanced fluorescence, and photochemical switching behavior in proteins.
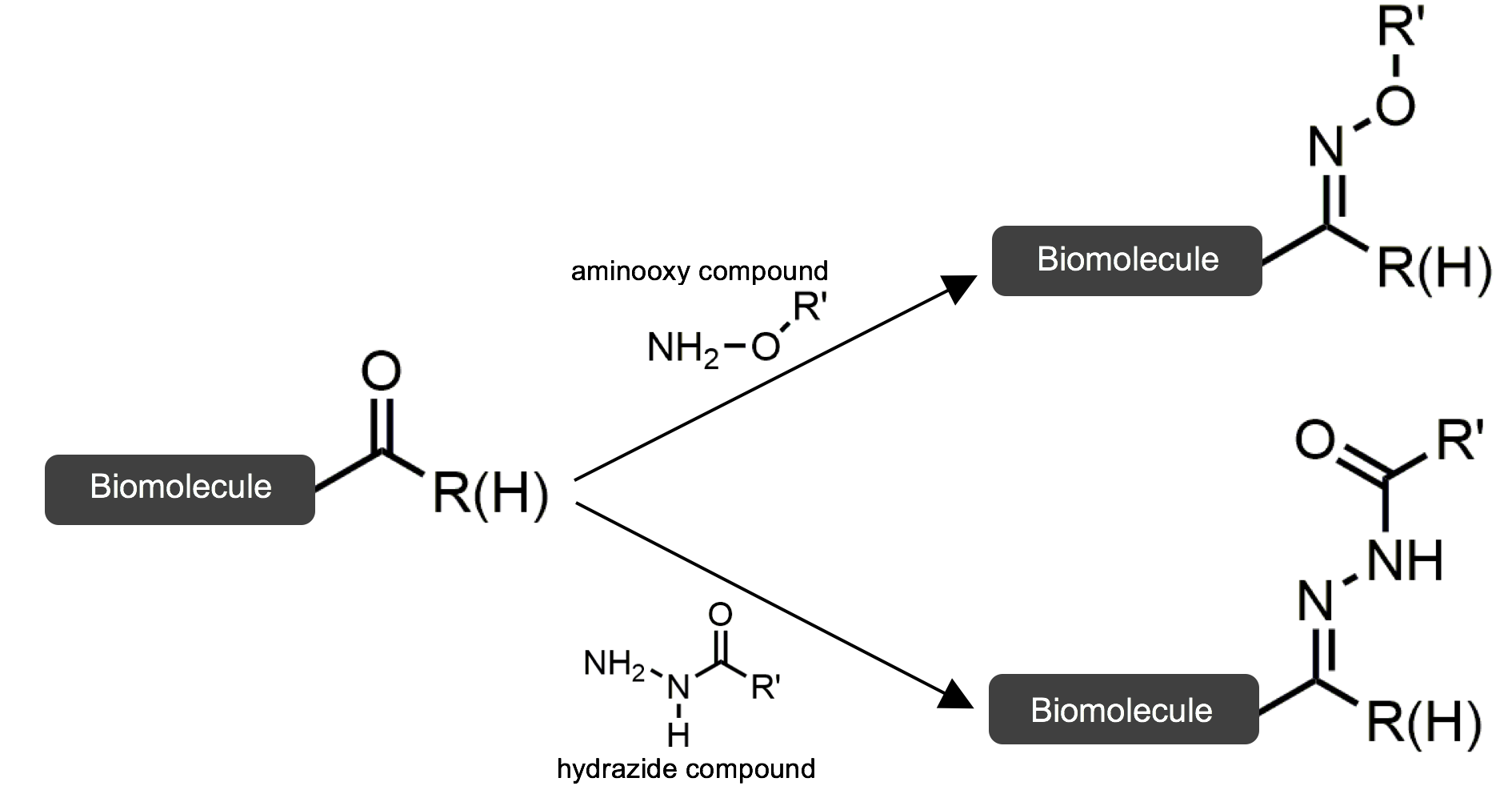
Ketone and aldehyde modification reactions are a common example of bioorthogonal reactions. These functional groups are effective because they are not present on cell surfaces and can thus be uniquely identified when attached. In this scenario, a ketone or aldehyde functional group on a biomolecule is coupled to a protein using aminooxy or hydrazide compounds to form stable oxime or hydrazone linkages between the biomolecule and protein. The figure below summarizes these reactions.
This reaction is efficient because the unique functional group is targeted. It also does not compete with innate processes.
N- and C- Termini Modification
Because natural amino acid residues are prevalent in proteins, it is difficult to selectively target a single residue. Therefore, techniques have been developed to target residues on the N- and C-termini because of enhanced site selectivity in these locations.
One example of N-terminal modification involves the oxidation of the serine or threonine residues to form an N-terminal aldehyde, which can undergo bioorthogonal reactions similar to the one described earlier.
An example of C-terminal modification is native chemical ligation (NCL), in which a thioester residue on the C-terminal is coupled to a thiol group on a cysteine residue on the N-terminal of another protein. This technique allows for the construction of large polypeptides by assembling smaller ones. NCL is especially powerful because the first step, the interaction between the thiol and thioester groups, is reversible, whereas the second step, which forms an amide, is irreversible. This leads to high yields of the final ligation product.
Labeling-Site Selectivity
The alkylation of cysteine residues is used to selectively modify a protein site. This technique is typically effective for naturally expressed proteins and also depends on the low abundance of cysteine residues. However, many of the reagents used in the alkylation process, such as iodoacetamides and vinyl sulfones, have been shown to modify other amino acids in the protein, which lowers selectivity.
Purification
Because most bioconjugation reactions do not go to full completion, excess reagent is often added. This makes it more difficult to extract the product out of the resulting mixture. In addition, very few general techniques have been developed to purify products of bioconjugation reactions. A simple technique such as size-exclusion chromatography can be used to isolate bioconjugated molecules if the product of interest is sufficiently large to elute faster than the other molecules present in the mixture. In other instances, purification methods unique to the bioconjugation reaction must be developed.
References
- Stephanopoulos, N. and Francis, M.B. (2011). "Choosing an Effective Protein Bioconjugation Strategy." Nature Chemical Biology. 7 (12): 876-84.
- Sletten, E.M. and Bertozzi, C.R. (2009). "Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality." Angew. Chem. Int. Ed. 48 (38): 6974-998.
- Carrico, I. S.; Carlson, B. L.; Bertozzi, C. R. (2007). "Introducing genetically encoded aldehydes into proteins". Nature Chemical Biology. 3 (6): 321–322.