28.3: Replication of DNA
- Page ID
- 36495
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)After completing this section, you should be able to describe, very briefly, the replication of DNA.
Make certain that you can define, and use in context, the key terms below.
- replication
- semiconservative replication
Notice that the objective for this section requires only that you be able to describe the replication process briefly.
According to the central dogma of molecular genetics, DNA is the genetically active component of the chromosomes of a cell. That is, DNA in the cell nucleus contains all the information necessary to control synthesis of the proteins, enzymes, and other molecules which are needed as that cell grows, carries on metabolism, and eventually reproduces. Thus when a cell divides, its DNA must pass on genetic information to both daughter cells. It must somehow be able to divide into duplicate copies. This process is called replication. Given the complementary double strands of DNA, it is relatively easy to see how DNA as a molecule is well structured for replication, as is show in Figure \(\PageIndex{1}\). Each strand serves as a template for a new strand. Thus, after DNA is replicated, each new DNA double helix will have one strand from the original DNA molecule, and one newly synthesized molecule. This is referred to as semiconservative replication.
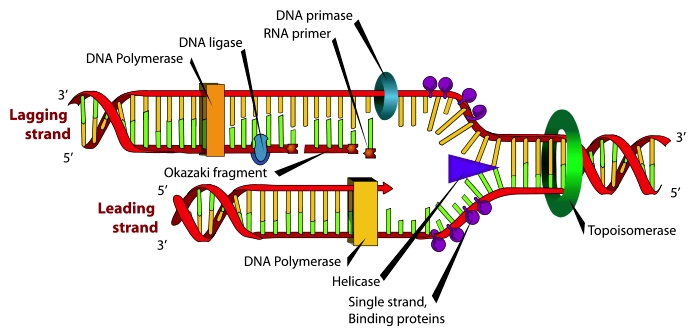
A rather complex mechanism exists for DNA replication, involving many different enzymes and protein factors. Let us consider some of the more important aspects of DNA replication. First, the double strand needs to be opened up to replicate each template strand. To do this, a set of proteins and enzymes bind to and open up the double helix at an origin point in the molecule. This forms replication forks, points where double stranded DNA opens up, allowing replication to occur. A helicase enzyme binds at the replication forks, with the function of further unwinding the DNA and allowing the replication fork to move along the double strand as DNA is replicated. Another enzyme, DNA gyrase, is also required to relieve stress on the duplex caused by unwinding the double strand. Further, single strand binding proteins are needed to prevent the single strands from reforming a double strand. Another essential enzyme in this initiation phase is primase, which creates an RNA primer on each single strand of DNA to begin replication from.
All of these initial functions are necessary to prepare the DNA for the main enzyme which builds then new strands, DNA polymerase. Multiple polymerase enzymes exist, but for the moment we will DNA polymerase III, the main DNA polymerase in E. coli. DNA polymerase III catalyzes the reaction by which a new nucleotide is added to a growing DNA strand. That reaction is seen in Figure \(\PageIndex{2}\). The DNA polymerase enzymes need a free 3' OH group in order to begin synthesizing a new strand, which explains the necessity of the RNA primer, which gives a 3'OH group for DNA polymerase III to start from.
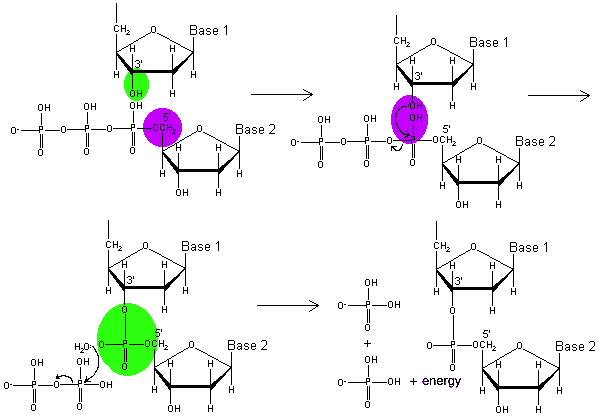
(Copyright; author via source)
This leads to another constraint on DNA polymerase III. One strand, the leading strand can be polymerized continuously since the new strand being created goes 5' to 3' from the replication fork, but since the original strands are anti-parallel, the other strand, the lagging strand is going in the wrong direction for polymerization. In this case, the polymerization reaction starts away from the replication fork and works back toward it. This means that the lagging strand is synthesized in disconnected segments, known as Okazaki fragments, instead of continuously. Later, another DNA polymerase, in the case of E. coli, DNA polymerase I, removes RNA primers and fills in the missing discontinuities. Then, another enzyme, DNA ligase, connects breaks between 3'OH groups and 5' phosphate groups in the newly synthesized strands that exist due to these discontinuities. While the enzymes of this process differ in eukaryotes, they fulfill similar mechanisms. Even with this complexity of this process, DNA polymerase III is able to add new nucleotides at a rate of 250-1,000 nucleotides per second.[3]
A number of advantages of the double-stranded structure held together by hydrogen bonds is evident in the process of replication. Complementary base pairing, A to T and G to C, insures that the two new DNA molecules will be the same as the original. The large number of hydrogen bonds, each of which is relatively weak, makes complete separation of the two strands unlikely, but one hydrogen bond, or even a few, can be broken rather easily. The helicase portion of the replication complex can therefore separate the two strands in much the same way that a zipper operates. Like the teeth of a zipper, hydrogen bonds provide great strength when all work together, but the proper tool can separate them one at a time.

The Scope of the Problem of DNA Replication
The 46 chromosomes of the human genome consists of roughly 6.5 billion base pairs of DNA if one considers the full diploid genome (i.e. if you count the DNA inherited from both parents). Considering that this DNA can be copied in just a few hours shows that the rate of DNA replication is staggering.
Although DNA replication is typically a highly accurate process and proofreading DNA polymerases help to keep the error rate low (about one per 10 billion bases), mistakes still occur. In addition to errors of replication, environmental damage may also occur to the DNA. Such uncorrected errors of replication or environmental DNA damage may lead to serious consequences. Therefore, Nature has evolved several mechanisms for detecting and repairing damaged or incorrectly synthesized DNA.
A segment of one strand from a DNA molecule has the sequence 5′-TCCATGAGTTGA-3′. What is the sequence of nucleotides in the opposite, or complementary, DNA chain?
Solution
Knowing that the two strands are antiparallel and that T base pairs with A, while C base pairs with G, the sequence of the complementary strand will be 3′-AGGTACTCAACT-5′ (can also be written as TCAACTCATGGA).
A segment of one strand from a DNA molecule has the sequence 5′-CCAGTGAATTGCCTAT-3′. What is the sequence of nucleotides in the opposite, or complementary, DNA chain?

