25.8: Structure and Function of Proteins
- Page ID
- 22364
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The biological functions of proteins are extremely diverse. Some act as hormones that regulate various metabolic processes. An example is insulin, which regulates blood-sugar levels. Enzymes act as catalysts for biological reactions, and other proteins serve as biological structural materials - for example, collagen and elastin in connective tissue and keratin in hair. Iron-containing proteins (hemoglobin and myoglobin in mammals) and copper-containing proteins (hemocyanins in shellfish) transport molecular oxygen. Some blood proteins form antibodies, which provide resistance to disease, while the so-called nucleoproteins are important constituents of the genes that supply and transmit genetic information in cell division. Motion by means of muscle contraction and the generation and transmission of nerve impulses also involve proteins.
How can a group of compounds, made from a common basis set of amino acids, be so remarkably heterogeneous and exhibit such varied yet specific functions? Clearly, the primary structure and presence or absence of special functional groups, metals, and so on, are of paramount importance. Of complementary importance are the three-dimensional structures of proteins, which are dictated not just by the primary structure but by the way the primary structure is put together biochemically. The polypeptide chains are seldom, if every fully extended, but are coiled and folded into more or less stable conformations. As a result, amino-acid side chains in distant positions in the linear sequence are brought into close proximity, and this juxtaposition often is crucial for the protein to fulfill its specific biological function.
Three-Dimensional Structure of Proteins
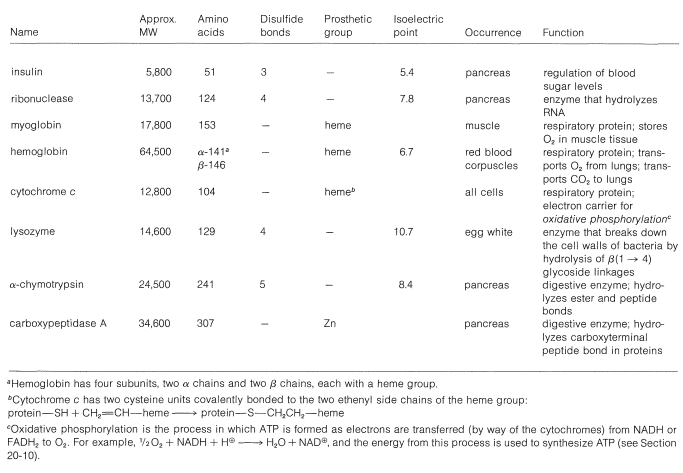
The elucidation of the detailed shape of protein molecules - in fact, the spatial locations of the individual atoms in a protein - is accomplished primarily by x-ray crystallography. The three-dimensional structures of more than twenty proteins have now been established by this technique. The importance of x-ray crystallography to structural and biological chemistry has been recognized in the award of six Nobel Prizes in this area.\(^6\) A number of important proteins and their properties are listed in Table 25-3.
Table 25-3: A Few Important Proteins of Known Structure
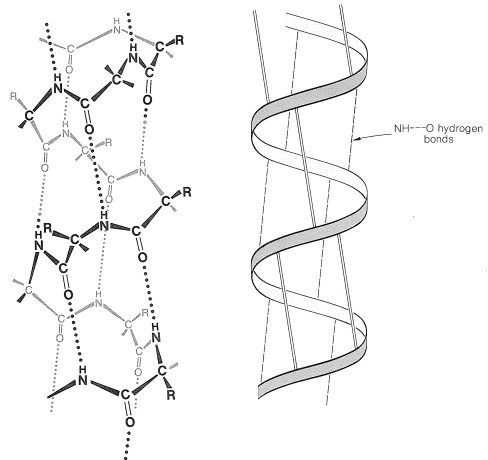
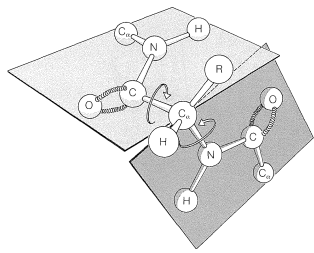
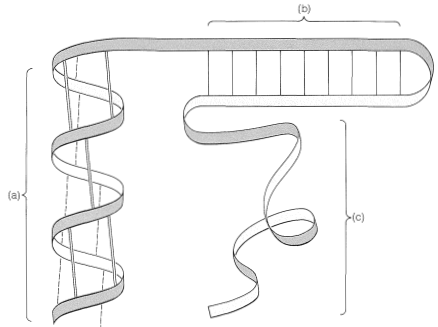
There are several other points to notice about the \(\alpha\) helix shown in Figure 25-11. The amide groups are planar and normally retain the stable trans configuration in the helical structure; bond lengths and bond angles are normal, and the \(\ce{-NH} \cdots \ce{O=C}-\) hydrogen bonds are nearly linear. However, the hydrogen bonds are not quite parallel to the long axis of the coil, so there are 3.6 rather than 4 amino-acid units per helical turn, and the spacing between turns is about \(5.4 \: \text{Å}\). The \(\alpha\) helix in proteins has right-handed turns like a right-hand screw thread.
The amino acids of the side chains lie outside the coil of the \(\alpha\) helix and are in close proximity to the side chains three and four amino-acid units apart. Because of this proximity, steric hindrance between large side chains can be sufficient to reduce the stability of the normal \(\alpha\) helix. When such hindrance occurs, there is a discontinuity in the helical structure, and the peptide chain may assume more random arrangements about the \(\ce{C-C}_\alpha\) and \(\ce{N-C}_\alpha\) bonds (see Figure 25-12), thereby allowing the molecule to fold back on itself and form new hydrogen bonds. The helical structure apparently is always interrupted at proline or hydroxyproline residues because the \(\ce{N-C}_\alpha\) bonds of these amino acids are not free to rotate (they are incorporated in five-membered rings) and also because the proline and hydroxyproline amide nitrogens have no hydrogens to participate in hydrogen bonding to carbonyl groups.
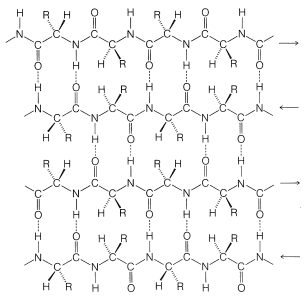
Pauling and Corey recognized a second stable conformation of polypeptide chains - the extended chain or \(\beta\)-pleated sheet (Figure 25-13). In this conformation the chains are fully extended with trans amide configurations. In this arrangement the distance is maximized between adjacent amino-acid side chains. Hydrogen bonding of the type \(\ce{-N-H} \cdots \ce{O=C}-\) is now between chains rather than between amino acids in a single chain (as in the \(\alpha\) helix). This type of structure is not as common as the \(\alpha\) helix and is found extensively only in silk fibroin. However, a number of proteins with a single polypeptide chain can form short sections of "antiparallel" \(\beta\)-pleated sheets by folding back on themselves, as illustrated in Figure 25-14.
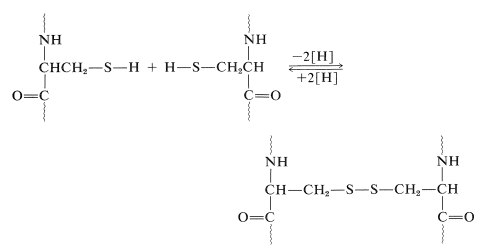
Another very important factor in protein architecture is the disulfide \(\ce{-S-S}-\) link. Remote parts of the polypeptide chain can be held close together through the oxidative coupling of two cysteine thiol groups to form a disulfide bridge:
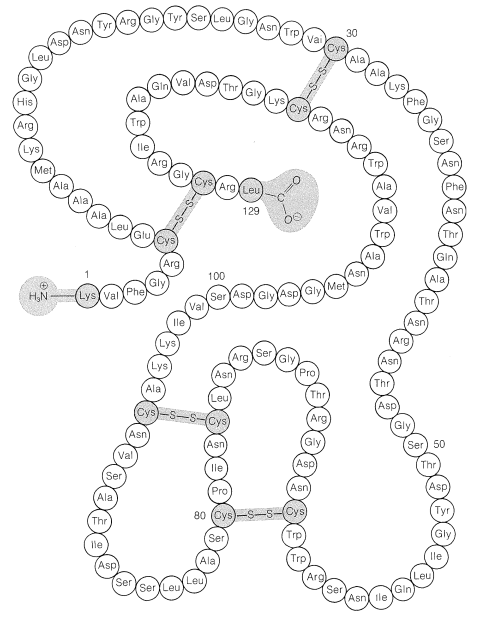
The disulfide bridges in some proteins are between different peptide chains. Insulin, for instance, has two interchain as well as one intrachain \(\ce{S-S}\) bridges (Figure 25-8).
Myoglobin and Hemoglobin
Some idea of the complexity of protein conformations can be gained from the structure of myoglobin. This protein is responsible for the storage and transport of molecular oxygen in the muscle tissue of mammals. It is a compact molecule of 153 amino-acid units in a chain that is extensively coiled as an \(\alpha\) helix. There are eight regions of discontinuity in the helical structure, and in these regions the chain folds on itself as shown in Figure 15-16. Four of the eight nonhelical regions occur at proline residues; the reason for the discontinuity at the other regions is not entirely clear. With the exception of two histidine units, the interior regions of myoglobin accommodate only the nonpolar side chains; the interior, therefore, is mostly hydrocarbonlike and repellant to water and other polar molecules. In contrast, the polar side chains are on the exterior of the protein.
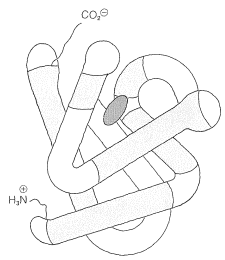
A number of proteins, including myoglobin, possess one or more nonpeptide components associated with specific sites on the polypeptide chain. These components are called prosthetic groups and are essential to the biological activity. When the prosthetic group is removed, the residual protein is referred to as an apoprotein.
In myoglobin the prosthetic group is a molecule of heme. The heme group belongs to a class of interesting compounds called metalloporphyrins, which are metal complexes of a highly conjugated ring system composed of four azacyclopentadiene (pyrrole) rings linked by \(\ce{-CH=}\) bridges between the 2 and 5 positions. The parent compound is known as porphin. Porphyrins have highly stabilized electronic excited states and absorb visible light. As a result they usually are brightly colored compounds (e.g., chlorophyll, Figure 20-6).
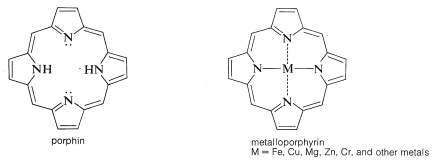
The porphyrin of heme is known as protoporphyrin IX, and the associated metal is iron [as \(\ce{Fe}\)(II) or \(\ce{Fe}\)(III)]. You will notice that the porphyrin ring carries methyl, ethenyl, and propanoic acid side chains:
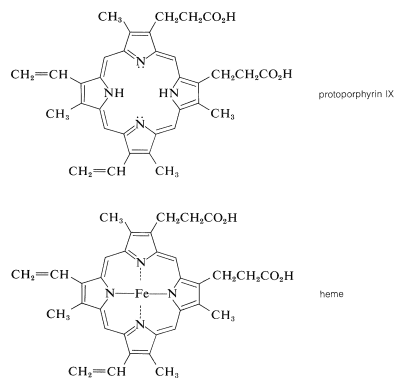
A major effort on the part of several eminent chemists in the early part of the century led to the elucidation of the structure of heme. The German chemist Hans Fischer successfully synthesized heme in 1929, a feat for which, in 1930, he received the Nobel Prize in chemistry. [Some years earlier (1915), Richard Willstatter received a Nobel Prize for structural studies of chlorophyll and plant pigments.]
A very important question is, how does the particular combination of protein and iron-porphyrin allow myoglobin to reversibly bind molecular oxygen? The answer to this question is not known in all its details, but it is well established that \(\ce{Fe}\)(II)-porphyrins will complex readily and reversibly with oxygen. There are two additional coordination sites around the iron in heme besides the four ring nitrogens. These are indicated below as the general ligands \(\ce{L}\):
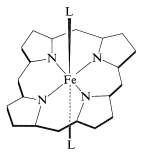
The disclike heme molecule fits into a cleft in the protein structure and is bound to it through one of the \(\ce{L}\) coordination sites to a histidine nitrogen. The remaining coordination site on the other side of the ring is occupied by molecular oxygen. In the absence of the coordination by histidine, the porphyrin iron would be oxidized rapidly to the ferric state, which does not bind oxygen.
A number of model compounds have been synthesized which have \(\ce{Fe}\)(II)-porphyrin rings carrying a side chain with histidine arranged to be able to coordinate with the metal on one side. Several of these substances show promise as oxygen carriers with properties similar to myoglobin.
Hemoglobin is related to myoglobin in both its structure and function. It reversibly binds molecular oxygen which it transports in the red corpuscles of blood rather than in muscle tissue. However, hemoglobin is made up of four polypeptide chains, in contrast to myoglobin which has only one chain. Two of the hemoglobin chains are of one kind with 141 amino acid residues, called the \(\alpha\) chains, and two are of another kind with 146 amino acids, called the \(\beta\) chains. Each chain, or subunit, contains one heme group identical with the heme in myoglobin. The subunits are held in the hemoglobin by noncovalent interactions and provide four hemes and hence four binding sites for molecular oxygen. The \(\alpha\) and \(\beta\) hemes have different affinities for oxygen but function in a cooperative way to increase oxygen availability to the cells.
In spite of the fact that the \(\alpha\) and \(\beta\) chains of hemoglobin are nonidentical with the myoglobin chain, the three-dimensional structures of all three chains are strikingly similar; myoglobins and hemoglobins differ slightly in amino acid composition, depending on the species but the protein shape remains essentially the same.
Quaternary Structures of Proteins
Many factors contribute to the three-dimensional structures of proteins. We already have mentioned hydrogen bonding between amide groups, location and character of prosthetic groups, and disulfide bonds. Other important influences include electrostatic interactions between ionic groups (\(\ce{-NH_3^+}\), \(\ce{-CO_2^-}\)), hydrogen-bonding involving side-chain substituents \(\left( \ce{-CH_2OH} \right)\), and nonbonded interactions. Except for the disulfide linkages, most of these interactions are weak compared to covalent bond strengths, and the conformations of many proteins can be altered rather easily. In fact, several have conformations that clearly are in dynamic equilibrium under physiological conditions. Such structural flexibility may be necessary for the protein to be functional, but if the conformation is altered irreversibly - that is, if it is denatured - its biological activity usually is destroyed.
In many cases there are important interactions between protein molecules that may lead to highly organized structures such as the pleated sheet of silk fibroin (Figure 25-13) or the coiling of \(\alpha\) helices, as found in \(\alpha\)-keratins, the fibrous proteins of hair, horn, and muscles (Figure 25-17). This sort of organization of protein molecules is called quaternary structure and is an important feature of many proteins that associate into dimers, tetramers, and so on. The tetrameric structure of hemoglobin is an important example.
\(^6\)The following Nobel laureates received their awards for contributions to the use of x-ray crystallography for structure determination: 1914, Max von Laue (physics), diffraction of x-rays in crystals; 1915, William Bragg and Lawrence Bragg (physics), study of crystal structure by means of x-rays; 1954, Linus Pauling (chemistry), study of structure of proteins; 1962, Max Perutz and John Kendrew (chemistry), structures of myoglobin and hemoglobin; 1962, Francis Crick, James Watson, and Maurice Wilkins (physiology and medicine), double helix of DNA; 1964, Dorothy Hodgkin (chemistry), determination of structure of vitamin B\(_{12}\) and penicillin by x-ray methods. She later determined the three-dimensional structure of insulin.
Contributors and Attributions
John D. Robert and Marjorie C. Caserio (1977) Basic Principles of Organic Chemistry, second edition. W. A. Benjamin, Inc. , Menlo Park, CA. ISBN 0-8053-8329-8. This content is copyrighted under the following conditions, "You are granted permission for individual, educational, research and non-commercial reproduction, distribution, display and performance of this work in any format."


