8.8: Genetic engineering
- Page ID
- 431342
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Understand recombinant DNA technology and its applications in biomedical technologies.
- Understand PCR and its applications in genetic testing, fingerprinting, and human genome project.
Recombinant DNA
DNA can be isolated from the cell and cut at a specific place using restriction enzymes. Fragments of DNA from different sources, e.g., an insulin gene from a human and a DNA from Escherichia coli (E. coli) cut using the same restriction enzyme, can be combined into a new recombinant DNA. The recombinant DNA can be introduced into the cell, e.g., back into E. coli, where it propagates using the cellular machinery of the host cell. It is then used to produce proteins of interest, e.g., human insulin used to treat diseases like diabetes.
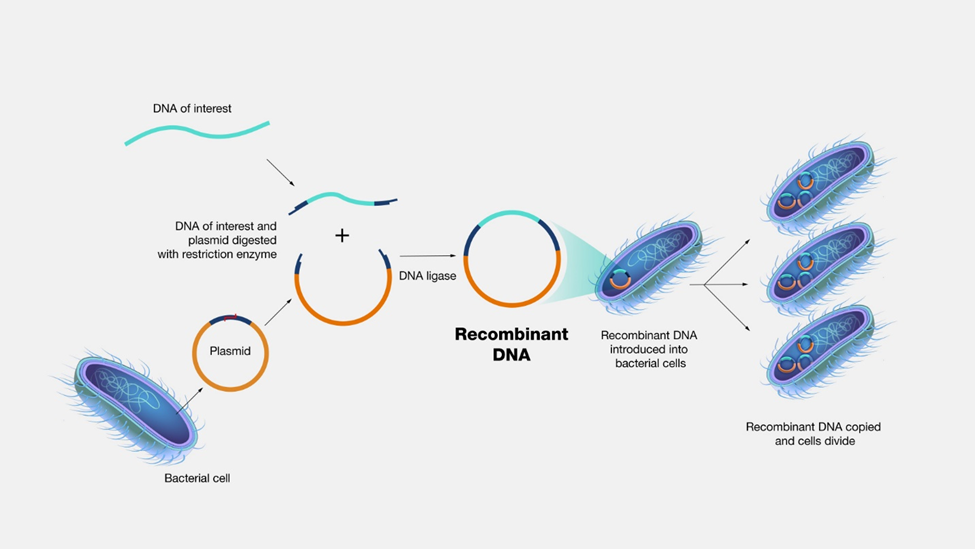
How is recombinant DNA produced?
The DNA of host cells, e.g., small circular plasmids of DNA from E. coli, are isolated first. The host cells are soaked in a detergent solution that disrupts the plasma membrane and releases the plasmids. A restriction enzyme is used to cut the DNA of the host cell at a specific location. The same enzyme is used to cut a piece of donor DNA, e.g., an insulin gene from a human. The cut ends of the DNA are called sticky ends. When the cut donor and host DNA are mixed, they join at the sticky ends, as illustrated in Figure \(\PageIndex{1}\). The resultant recombinant DNA is introduced to a new culture of E. Coli. The host cells that have taken up the recombinant DNA are selected and grown. When these bacteria grow and divide, they produce the protein encoded in the inserted gene, i.e., insulin in this example. Insulin is extracted, purified, and used to treat diabetes.
The recombinant DNA technology is used in food production, medicine, agriculture, and bioengineering. Some examples are listed below.
- Chymosin is an enzyme used to manufacture chees. Currently, ~60% of hard cheese is produced in the US using genetically engineered chymosin.
- Insulin is usually synthesized by inserting the human insulin gene into E. Coli, or yeast, produces insulin processed and used to treat diabetes.
- Human growth hormone (HGT) is needed for patients whose pituitary glands do not produce sufficient hormones. Recombinant HGT is commonly used for this purpose.
- Blood clotting factor VIII is needed for patients suffering from the bleeding disorder hemophilia. It is produced these days by recombinant DNA technology.
- The Hepatitis B vaccine needed to treat hepatitis B infection is produced by recombinant DNA technology.
- The recombinant HIV protein tests the presence of antibodies that may have been produced in response to an HIV infection.
- Herbicide-resistant crops, including soy, corn, canola, alfalfa, cotton, etc., have been developed that contain recombinant genes resistant to the herbicide glyphosate found in Roundup. It allowed the use of Roundup to control weeds without affecting the crop.
- Insect-resistant corps: Bacillus thuringeiensis is a bacteria that naturally produces a protein with insecticidal properties. Crops containing the recombinant gene from the bacteria hold promise to control insect predators without using an insecticide.
- Interferon produced by recombinant DNA technology is used to treat cancer and viral disease.
- The influenza vaccine is used to prevent influenza.
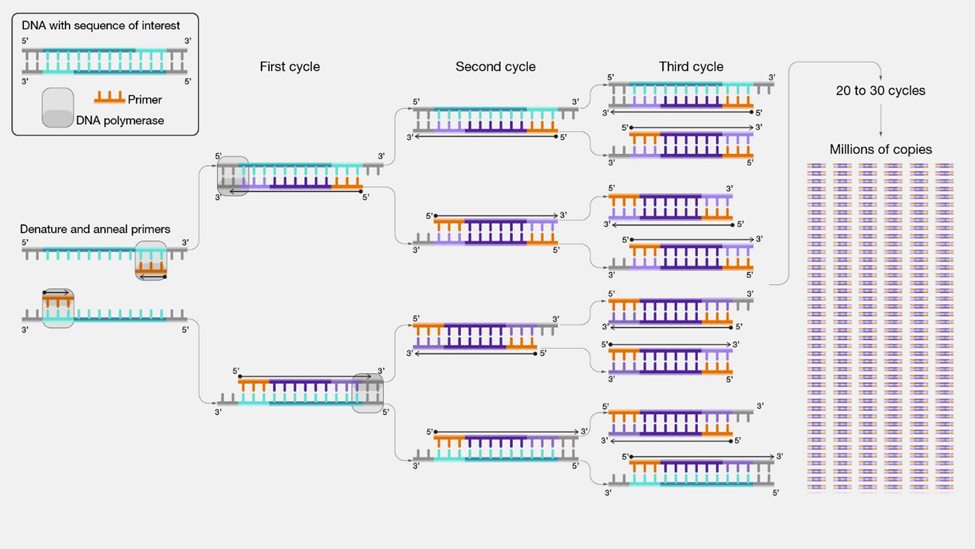
Polymerase chain reaction (PCR)
polymerase chain reaction (PCR) quickly produces millions of copies of a DNA segment of interest. The selected DNA is heated to denature and separate the two strands and mixed with DNA polymerase, a short synthetic DNA called primer to select the segment to be amplified, and nucleotides, to produce a complementary strand. The process is repeated several times to make millions of copies of the desired DNA fragment, as illustrated in Figure \(\PageIndex{2}\).
Genetic testing
Genetic testing is done in a laboratory to study an individual's DNA to diagnose a defective gene that may cause a congenital disease, ancestry studies, or forensic studies. For example, breast cancer may be related to defects in breast cancer genes BRCA1 and BRCA2. Patients are screened for these defects by using blood or saliva samples to extract the DNA, and the PCR technique is applied to amplify and study the defective genes. Genetic testing is also used to study the DNA of tumors or cancer cases.
Fingerprinting
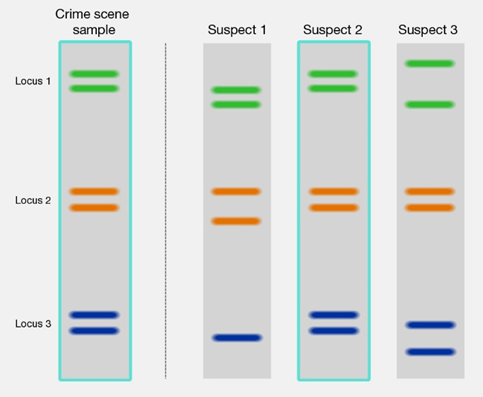
Fingerprinting is a technique based on PCR that allows determining the nucleotide sequence of some areas of human DNA that are unique to individuals. A small sample from flood, skin, saliva, or semen is used to extract the DNA for amplification by PCR. Fluorescent or radioactive isotopes are incorporated in the amplified DNA for easy monitoring. The DNA is cut into pieces by restriction enzymes, placed on a gel, separated by electrophoresis, and studied to identify the individual, as illustrated in Figure \(\PageIndex{3}\). This technique is used for paternity tests, criminal investigations, and forensics.
Human genome
The human genome project meant to study all human DNA (human genome) comprehensively, was launched in October 1990 and completed in April 2003. It shows about 20,000 protein-coding genes in humans, representing only about 1.5% of the human genome. The role of the rest of the genome is still being explored. It is mainly related to regulating genes and serving as a protein recognition site. These studies provide fundamental information to study human biology, defects in DNA that lead to genetic diseases, and find cures for conditions.


