7.2: Complex I
- Page ID
- 190046
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Complex I is a collection of proteins that serves as one of two entry points into oxidative phosphorylation; the other is complex II. Both of these complexes accept electrons from molecules produced in the catabolism of glucose. By shuttling these electrons from one electron acceptor to another, generally moving to higher potential (remember, in redox terminology, that means lower energy) the assembly of proteins that take part in oxidative phosphorylation are able to produce ATP. ATP, in turn, is used to power other metabolic processes.
The main events in Complex I are summarised in the cartoon below. You can see the electrons entering from the matrix at the bottom of the picture (the pathway is shown by the blue arrows). They are delivered by NADH and handed off to FMN; this step will be discussed below. The electrons are transferred via outer sphere electron transfer through a series of iron sulfur clusters and are eventually delivered to the lipid-soluble ubiquinone (Q).
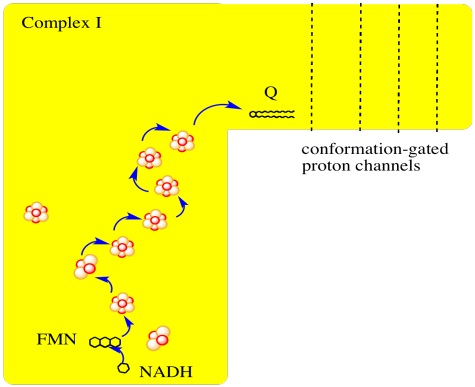
- Complex I marks the beginning of the electron transport chain.
- Electrons are delivered from NADH, pass through Complex I, and finally reach a ubiquinone.
- The energy released during electron transport helps pump protons across the complex.
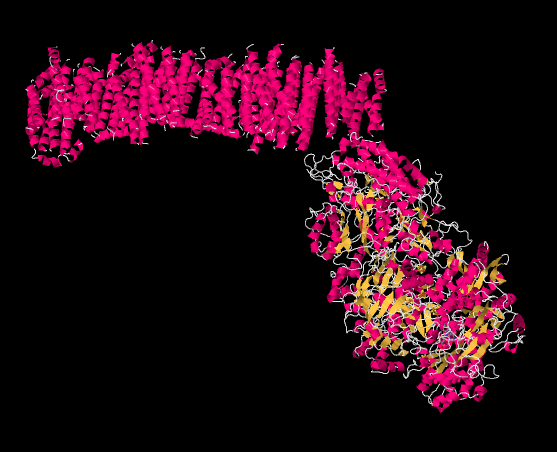
The picture below is from an X-ray crystal structure of Complex I (the source of the data is cited at the end of this page). Rather than showing every atom, which is the usual result of a crystal structure, the data are displayed in a "cartoon" form, so that you can get a better sense of the overall structure. The picture is also colour-coded to help you see structures more clearly. The pink helices (spirals) along the top are the portion of the complex that is bound in the inner mitochondrial membrane. The membrane, too, would extend acorss the top of the picture from left to right. It is easy to imagine the parallel bundle of α-helices fitting nicely in the midst of the parallel array of phospholipids that form the membrane. The egg-shaped, yellow and pink form to the lower right is the part of the complex that extends out into the mitochondrial matrix. The yellow portions indicate β-sheets, whereas the white threads indicate loops.
Exercise \(\PageIndex{1}\)
Specific amino acids are likely to be found along the α-helices that bind in the membrane. Indicate different possibilities below.

- Answer
-

In the case of Complex I, the electrons are introduced in the form of NADH. Electrons travel through Complex I and eventually are delivered to ubiquinone; ubiquinone carries the electrons to the next stage of the oxidative phosphorylation supercomplex, which is Complex III. Because Complex I takes electrons from NADH and delivers them to ubiquinone, Complex I is also referred to as "NADH:ubiquinone oxidoreductase".
NADH is produced during glycolysis and the TCA cycle. Remember, NADH is a two-electron donor: it donates a hydride ion to a substrate, becoming NAD+. A hydride ion, of course, is just a proton and two electrons.
- NADH produced in glycolsis and the TCA cycle delivers a pair of electrons to Complex I.
- The NADH is delivered to complex I from the mitochondrial matrix (in the interior of the mitochondrion).
By far the most common electron acceptor in the oxidative phosphorylation supercomplex is an iron atom. Of course, the most common oxidation states for iron ions are Fe2+ and Fe3+. An iron in the 3+ oxidation state is able to accept an electron, becoming Fe2+. In contrast, an iron in the 2+ oxidation state could pass an electron on, becoming Fe3+ in the process.
Picture a bucket brigade, in which people passing buckets of water from one to another all act together to put out a fire. The iron atoms act in much the same way, each passing an electron to the next in order to complete the electron transport chain.
- Electron transport is accomplished via many small steps rather than a few large ones.
If we strip away the proteins from Complex I, we can get a picture of some of the other pieces inside. Looking at the X-ray crystal structure data, we can simply ignore every atom in the protein, until we are left with the "ligands". In biochemistry, ligands means the stuff attached to the proteins (as opposed to in inorganic chemistry, where it means the stuff attached to the metals). That's what we see below. The red and yellow shapes that you see are iron sulfide complexes, strung along so that they can pass electrons along through Complex I. The pieces that we see here are found within the hydrophilic part of Complex I; that is, they are found within the yellow and pink egg-shaped part seen in the structure above. This picture is oriented in the same direction as the one above; the iron complexes extend from the lower part of the hydrophilic domain all the way up to the edge of the hydrophobic, membrane-bound portion of the complex.

So, we have a "wire" to carry the electrons through the complex after they are delivered by NADH. We have a mismatch problem, however. NADH is a two electron donor. An Fe3+ ion is a one-electron acceptor. We need an adapter for this electrical connection. The adapter comes in the form of FMN. FMN is the structure with some atoms coloured in blue and red near the bottom right corner of the picture.
- NADH only donates two electrons at a time.
- The iron ions in the electron transport chain can toggle between Fe(III) and Fe(II); they can accept only one electron at a time.
- An adapter is needed to convert two-electron transfer into one-electron transfer.
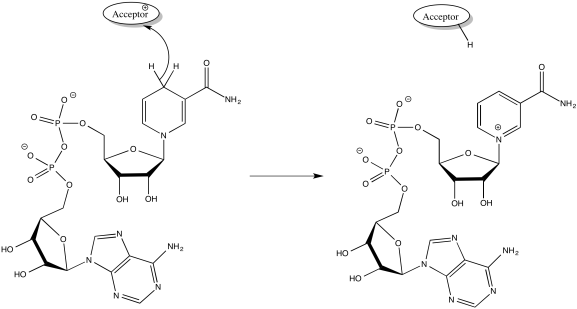
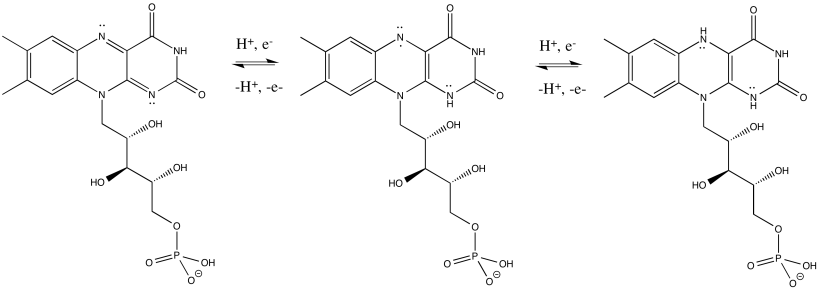
FMNH2 is a little bit like NADH. Its oxidized form, FMN, can accept two electrons and a proton in the form of a hydride ion, as well as an additional proton. In other words, FMN accepts H- and H+ to become FMNH2. However, a slightly different route is available to FMN. It can also undergo reduction one electron at a time. In reality, the addition of an electron would be shortly preceded by or shortly followed by addition of a proton, in order to keep the overall charge the same. This state, FMNH, is called the semiquinone form.
What's the difference between NAD and FMN? Why is one able to accept only a pair of electrons, whereas the other can accept one at a time? When FMN accepts one electron, it becomes a radical. Radicals are unstable, reactive species. They can be stabilised chiefly by delocalisation. The additional conjugation in FMN compared to NAD allows the odd electron to be delocalised more extensively in FMN. That radical stability is the key difference.
- The presence of extended conjugation stabilises a radical on FMNH.
- The stability of this radical allows FMN to accept one electron at a time.
Exercise \(\PageIndex{2}\)
Provide a mechanism for the conversion of FMN to FMNH2 in the presence of NADH and a lysine side chain, at pH 7.
- Answer
-
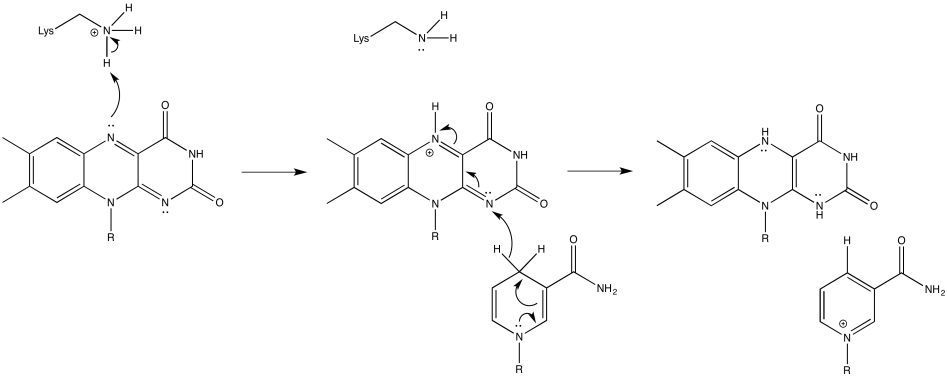
Once FMNH2 has formed, the reverse is true, of course. It can give up one electron at a time. As a result, FMN can take a pair of electrons coming in from NADH and send them out one at a time into the electron transport chain.
The rest of the electron transport chain through Complex I is a series of iron-sulfur clusters. As the name suggests, these clusters consist of iron and sulfur atoms. The most common variety contains four iron atoms and four sulfur atoms arranged at the corners of a cube. These clusters are often referred to as 4Fe4S clusters for obvious reasons: there are 4 iron atoms and 4 sulfur atoms. The sulfur atoms at the corners are really sulfide ions, S2-. In addition to the three sulfur ions, each of the iron atoms is also bound to an anionic cysteine residue, so that the iron has tetrahedral coordination geometry. The iron atoms are present as a combination of Fe2+ and Fe3+ ions.
- Iron sulfur clusters are very common in biological electron transport.
- The iron ions can be Fe(II) or Fe(III).
- The ligands for the iron ions include sulfide ions, S2-.
- The iron sulfide clusters are usually held in place in the protein by cysteine ligands (CysS-).
Here is another view of the ligands, seen from a different point of view this time. See whether you can find a group of four iron atoms (coloured red) bound with four sulfur atoms (coloured yellow).
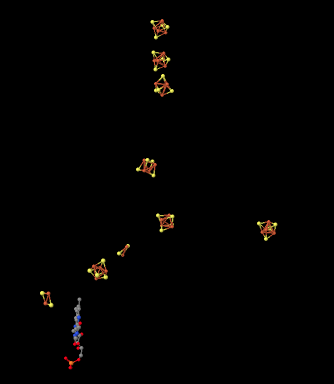
There are other variations of FeS clusters. A very common one is a 2Fe2S cluster, which of course consists of two iron atoms and two sulfide ions at alternating corners of a diamond. There are a couple of these clusters visible in the picture above. Again, the iron atoms could be Fe2+ or Fe3+ ions, or one of each. Also, each iron is usually bound to two additional cysteine anions to complete a tetrahedral geometry. Those groups aren't shown here, though, because we have left the protein out of the picture. (We will see eventually that other amino acids occasionally bind iron sulfur clusters in place of cysteine.)
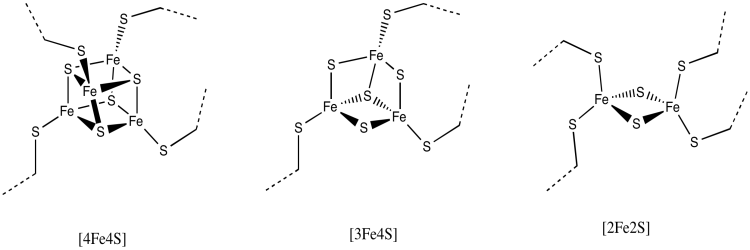
Another possibility is a 3Fe4S cluster, a lop-sided affair in which one of the iron atoms is essentially left out of the FeS cube.
Exercise \(\PageIndex{3}\)
Assuming one iron is present as Fe(III) and the rest as Fe(II), calculate the overall charges on:
a) 2Fe2S b) 3Fe4S c) 4Fe4S
- Answer a
-
a) Iron charges: Fe(II) + Fe(III) = 5+
Ligand charges: 2 sulfides = 2 x 2- = 4- ; 4 cysteines = 4 x 1- = 4- ; total = 8-
Overall: 3-
- Answer b
-
b) Iron charges: 2 x Fe(II) + Fe(III) = 4+ + 3+ = 7+
Ligand charges: 4 sulfides = 4 x 2- = 8- ; 3 cysteines = 3 x 1- = 3- ; total = 11-
Overall: 4-
- Answer c
-
c) Iron charges: 3 x Fe(II) + Fe(III) = 6+ + 3+ = 9+
Ligand charges: 4 sulfides = 4 x 2- = 8- ; 4 cysteines = 4 x 1- = 4- ; total = 12-
Overall: 3-
Exercise \(\PageIndex{4}\)
Environment plays a role in modulating reduction potentials in proteins. Suppose a 2Fe2S cluster was in a mixed Fe(II)/(III) oxidation state. How would its reduction potential when surrounded by nonpolar amino acid residues compare to its reduction potential when surrounded by polar amino acid residues?
- Answer
-
Upon reduction, the charge on a 2Fe2S cluster will increase from 3- to 4-, assuming it starts in a mixed Fe(II)/(III) state (whereas if it starts in a Fe(III)/(III) state, the overall charge will increase from 2- to 3-). These anions would be stabilised by strong intermolecular interactions such as ion-dipole forces. Both states (oxidised and reduced) will be stabilised by a polar environment, but the more highly charged reduced state will depend even more strongly on stabilisation by the environment. As a result, we might expect the reduction potential to be lower when surrounded by nonpolar amino acid residues, and higher if surrounded by polar residues.
There is a whole series of these clusters in Complex I. The electrons delivered from NADH are sent from one to the next and then to the next. There are a couple of reasons for this arrangement.
Because these clusters are all bound in place by the protein -- completely immobilised -- the electron transfer has to occur via an outer sphere mechanism. Why are there so many clusters? Remember, outer sphere electron transfers have a limited range. The electron, under most circumstances, can only hop so far. By providing a series of conducting FeS clusters, the electron can hop from one to the next, arranged like stepping stones across a river.
Not only that, but the electron transport chain takes electrons from NADH and delivers them, ultimately, to molecular oxygen in Complex IV. The molecular oxygen is converted to water. That transfer from NADH to water is very exothermic. The reaction is very much downhill in energy. In order to make this transfer practically feasible, and in order to harness the enormous amount of energy involved, the electrons are allowed to step downhill just a little at a time.
Occasionally, electrons may even hop back uphill slightly, recapturing energy that has been lost to the surroundings in previous transfers. This damping effect may make the whole process more efficient. Still, the electrons are overall rolling downhill energetically. An electron may jump uphill a few times, but eventually those uphill jumps will be followed by a downhill drop, so that overall the electron has moved to lower energy.
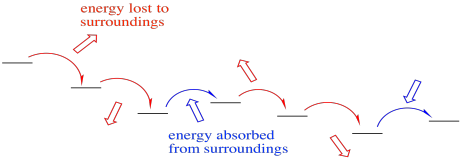
- An occasional uphill electron transfer absorbs energy.
- Re-absorption boosts efficiency by preventing heat loss.
In Complex I, the final destination for the electron transport chain is a ubiquinone, sometimes abbreviated as Q or UQ. Like FMN, UQ is a two electron, two proton acceptor, to become UQH2. Also like FMN, UQ can accept one proton and one electron at a time, to form the semiquinone form, UQH. Once again, this is a radical species.
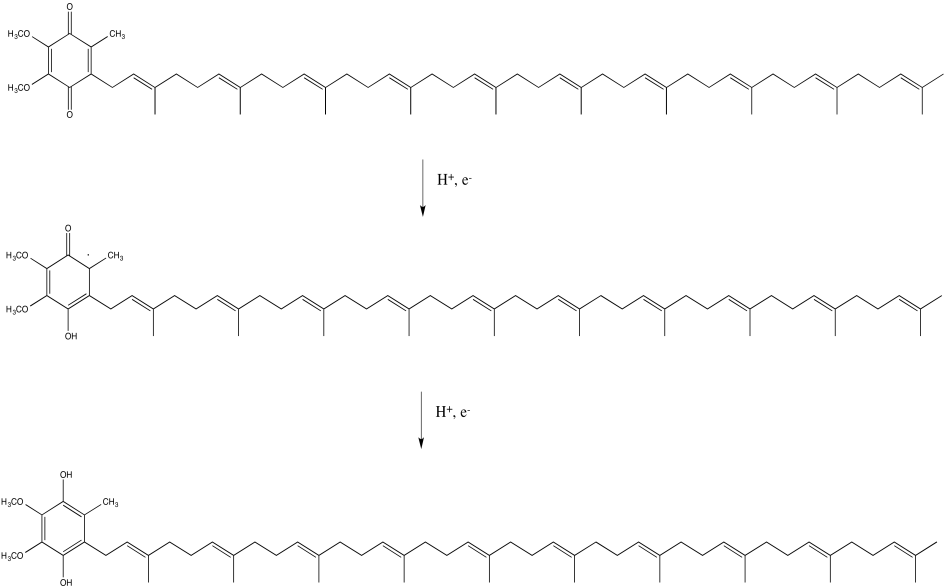
Exercise \(\PageIndex{5}\)
Provide a mechanism for the conversion of UQ to UQH2 in the presence of Fe2+ ions and lysine side chains, at pH 7.
- Answer
-
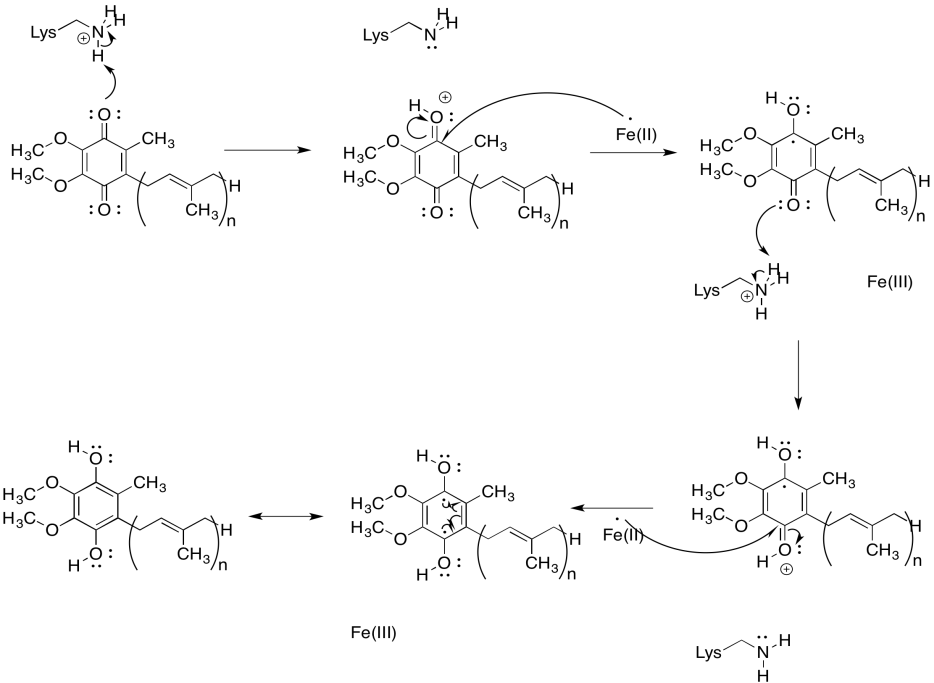
UQ is very different from the FeS clusters or the FMN because it isn't attached to a protein. It isn't tied down. It can move around. That makes it a mobile electron carrier. In addition to being a relatively high-potential electron acceptor (at least compared to other things in Complex I), the role of UQH2 is to deliver electrons to Complex III so that the electron transport chain can continue.
The trouble is, if UQH2 is mobile, what's to keep it from wandering away? How is its pathway limited so that it is more likely to reach its destination? Remember that the oxidative phosphorylation supercomplex is a group of membrane-bound proteins. They are held in a lipid-rich environment. The structure of UQH2, with its long tail, is very lipophilic. If it stays in the membrane, its movements are limited to two dimensions, rather than three, and it is much more likely to reach its destination of Complex III.
- Ubiquinone is a lipid-soluble, mobile electron carrier.
- Its movement is restricted to the mitochondrial membrane.
There is one more important feature of Complex I. Like some of the other complexes involved in oxidative phosphorylation (Complexes III and IV), Complex I pumps protons across the inner mitochondrial membrane. Ultimately, the protons that have gathered on the edge of the inner mitochondrial membrane cascade back across, turning a molecular millwheel that drives the manufacture of ATP. The ATP is used to power processes throughout the cell.
- Protons are actively pumped across the mitochondrial membrane.
- As a result, a charge develops across the membrane.
- The mitochondrial matrix becomes "n-doped" or negatively charged.
- The intermembrane space becomes "p-doped" or positively charged.
This proton pump is an example of active transport. Energy is expended to transport protons across the membrane, despite a buildup of positive charge in the intermembrane space (and a corresponding buildup of negative charge in the matrix). The energy released by the electron transport chain may be responsible for conformational changes in the protein that help this transport occur.
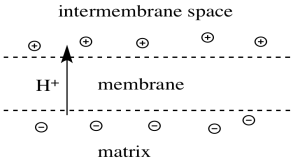
There still seems to be some discussion going on about how, exactly, the proton pump works in this system. However, some things are clear. The transport of proteins across a hydrophobic membrane is likely facilitated by the presence of hydrophilic portions of the protein. There are believed to be channels that open up in the protein, allowing water molecules to move through the protein. Because the protein is embedded in the membrane, these protons are also crossing the membrane at the same time.

Exercise \(\PageIndex{6}\)
Frequently, specific amino acids can play a role in assisting the transport of protons (or other ions). Fill in some different possibilities for these amino acids.

- Answer
-

Exercise \(\PageIndex{7}\)
In general, the proton that enters the complex on one side of the membrane is probably not the same proton that emerges on the other side. Provide a mechanism with arrows to illustrate this process.
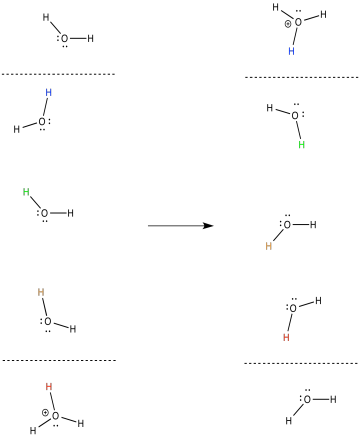
- Answer
-

The release of energy over the electron transport chain drives the transport of protons across the membrane. There is another factor that helps, too. Like the electrons, the protons travel from the matrix toward the intermembrane space. Both positive and negative chages are traveling in the same direction. That raises the possibility of coupled transport, in which the flow of electrons through the protein makes it easier for protons to follow along (or vice versa).
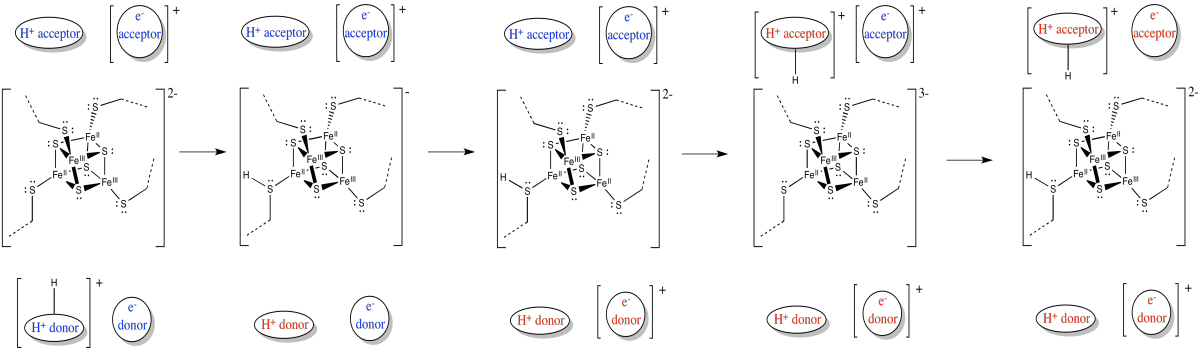
In coupled transport, the movement of an proton is quickly followed by the transfer of an electron (or vice versa).
- Electrons and protons are travelling in the same direction through complex I: from the matrix toward the intermembrane space.
- Their opposite charges may lead to a coupled mechanism in which the movement of one makes it easier for the other to follow.
Complex I is not the only entry point for electrons into the electron transport chain. Complex II plays a similar role. Together, they harvest energy from the electron transport chain; that energy is ultimately used to make ATP, which can move through the cell to release energy elsewhere.
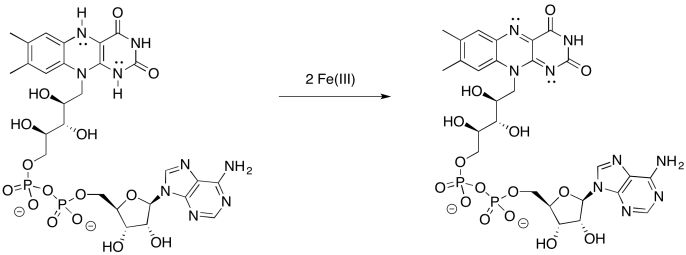
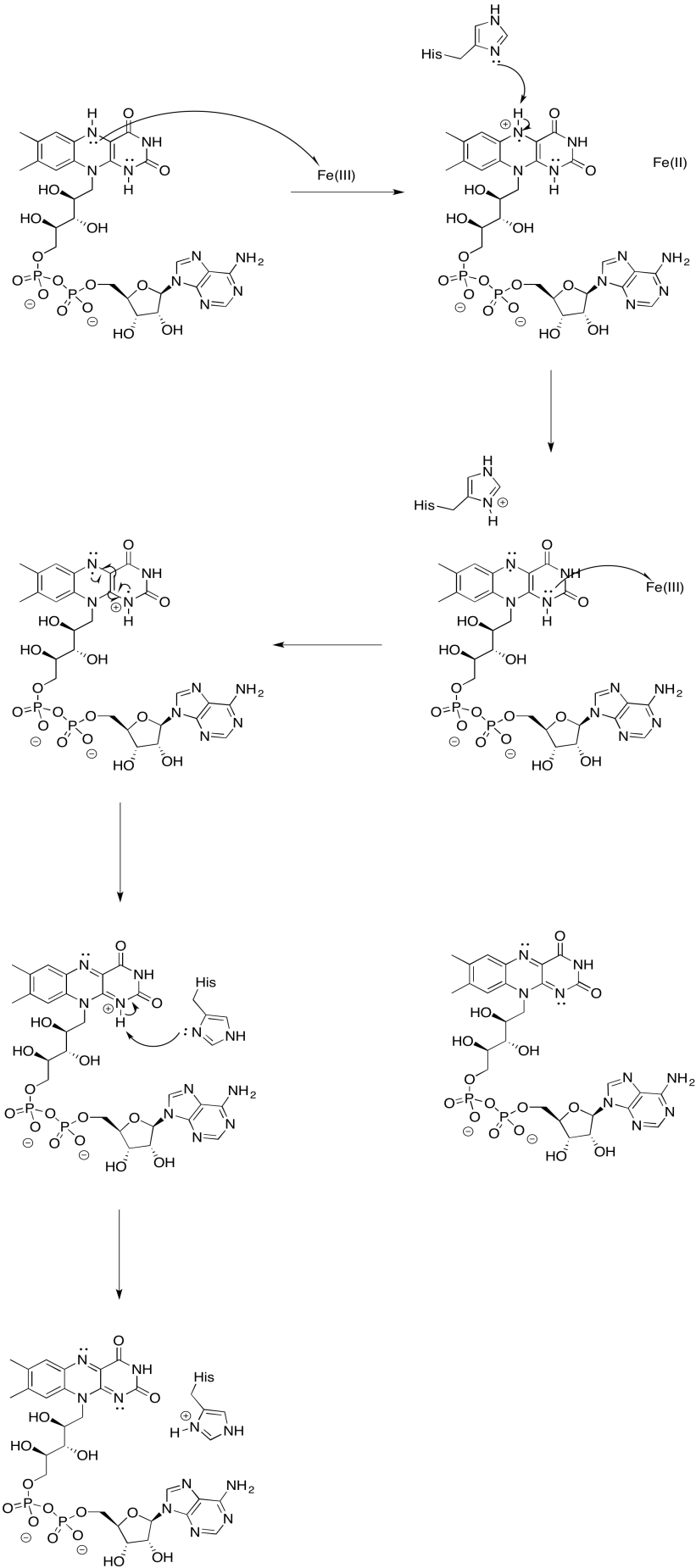
Exercise \(\PageIndex{8}\)
Provide a mechanism for the oxidation of FMNH2 by iron(III).
- Answer
-
Exercise \(\PageIndex{9}\)
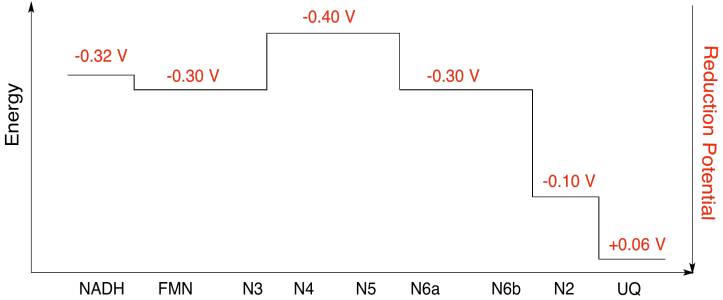
It's difficult to measure the reduction potential of an individual site within a protein. However, researchers have been able to estimate these values by measuring EPR spectra under various conditions. For example, here is an approximate picture of potentials in Complex I.
- Two of the N clusters are probably not directly involved in the electron transport chain. Which ones?
- Use the data in the diagram to construct a potential energy diagram for the transfer of the electron along the pathway.
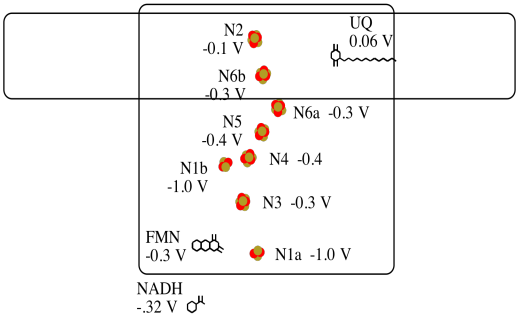
- Answer a
-
a) N1a and N1b are most likely not involved, because their reduction potentials are too negative.
- Answer b
-
b)
Exercise \(\PageIndex{10}\)
Using the values in the figure above, calculate the energy change when an electron is transferred from the N5 cluster to the N6a cluster.
- Answer
-
Assuming the reduction potentials are:
N5(ox) + e- → N5(red) Eored = -0.40 V
N6a(ox) + e- → N6a(red) Eored = -0.30 V
Then the potential difference for the reaction, ΔEo = -0.30 - (-0.40) V = 0.10 V.
The Faraday relation ΔG = - n F ΔEo gives
ΔG = - 1 x 96,485 J V-1 mol-1 x 0.10 V = 9,649 J mol-1 = 9.7 kJ mol-1


