2.4: Epigenetic Mechanisms
- Page ID
- 308330
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- 1: Identify the main epigenetic mechanisms related to control of gene expression.
4.1
Introduction
 Epigenetics is defined as potentially heritable and reversible changes in gene expression mediated by methylation of DNA, modifications of histone proteins or by non-coding RNAs that are not due to any alteration in the DNA sequence. These processes singularly or jointly affect transcript stability, DNA folding, nucleosome positioning, chromatin compaction, and ultimately nuclear organization. They determine whether a gene is silenced or activated and when and where this occurs.
Epigenetics is defined as potentially heritable and reversible changes in gene expression mediated by methylation of DNA, modifications of histone proteins or by non-coding RNAs that are not due to any alteration in the DNA sequence. These processes singularly or jointly affect transcript stability, DNA folding, nucleosome positioning, chromatin compaction, and ultimately nuclear organization. They determine whether a gene is silenced or activated and when and where this occurs.
Epigenetic change is a regular and natural occurrence, essential for normal cell development, but can also be influenced by several factors including age, the environment/lifestyle, and disease state.
4.2
DNA Methylation
 DNA methylation is a covalent modification of DNA, in which a methyl group is transferred from S-adenosylmethionine (SAM), that is converted to S-adenosylhomocisteine (SAH), to the 5 position of cytosine by a family of enzymes known as DNA methyltransferases (DNMT).
DNA methylation is a covalent modification of DNA, in which a methyl group is transferred from S-adenosylmethionine (SAM), that is converted to S-adenosylhomocisteine (SAH), to the 5 position of cytosine by a family of enzymes known as DNA methyltransferases (DNMT).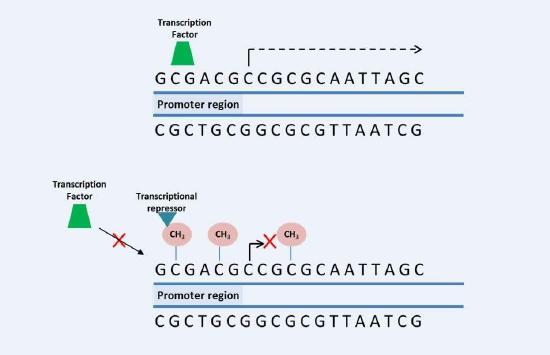 DNA methylation occurs predominantly in cytosines located in proximity of guanines, known as CpG dinucleotides (CpGs) or CpG island. These CpG island are found in promoter region of genes. The methyl group inhibits the binding of transcription factors to their recognition site, resulting in inhibition of gene transcription. Furthermore, the methyl group may attract other proteins know as transcriptional repressors that contributes for inhibition of gene transcription.
DNA methylation occurs predominantly in cytosines located in proximity of guanines, known as CpG dinucleotides (CpGs) or CpG island. These CpG island are found in promoter region of genes. The methyl group inhibits the binding of transcription factors to their recognition site, resulting in inhibition of gene transcription. Furthermore, the methyl group may attract other proteins know as transcriptional repressors that contributes for inhibition of gene transcription.4.3
Histones Post-Translational Modifications
The nucleosome is composed of five histone proteins (H1, H2A, B, H3, and H4). The N-terminus of these histone proteins are subject to covalent modifications such as methylation, phosphorylation, acetylation, ubiquitination or sumoylation by a group of histone-modifying enzymes . Alterations in these proteins contribute to the accessibility and compactness of the chromatin, and result in activation or suppression of particular genes.
EXAMPLES OF TYPES AND ROLES OF HISTONE MODIFICATIONS

Activation of gene Transcription:
Acelylation in the lysine 9(K9) of histone 3(H3) - H3K9ac
Acetylation in the lysine 27(K27) of histone 3(H3) - H3K27ac
Trimethylation in the lysine 4(K4) of histone 3(H3) - H3K4me3
Repression of gene transcription:
Trimethylation in the lysine 4(K3) of histone 3(H3) - H3K9me3
Trimethylation in the lysine 27(K27) of histone 3(H3) - H3K27me3
Trimethylation in the lysine 20(K20) of histone 3(H3) - H3K20me3
 DNA methylation and histone post-translational modifications play important role in the establishment of chromatin structure and in consequence in the gene expression modulation. These epigenetic mechanisms can be independent or can happen together to control the gene expression. They also may influence each other, for example the histone methylation can help to direct DNA methylation patterns, and DNA methylation might serve as a template for some histone modifications after DNA replication.
DNA methylation and histone post-translational modifications play important role in the establishment of chromatin structure and in consequence in the gene expression modulation. These epigenetic mechanisms can be independent or can happen together to control the gene expression. They also may influence each other, for example the histone methylation can help to direct DNA methylation patterns, and DNA methylation might serve as a template for some histone modifications after DNA replication.4.4
microRNAs
miRNAs have an important role in gene regulation and they can influence biological functions including cell differentiation and proliferation during normal development and pathological responses. They are small non-coding RNA molecules (containing about 22 nucleotides), derived from regions of RNA transcripts that fold back on themselves to form short hairpins. They regulate gene expression at the post-transcriptional level. A number of miRNAs may bind to specific regions of the messenger RNA (mRNA) and block its translation to proteins. Alteration of the expression of miRNAs is believed to contribute to the progression of tumorigenesis and other diseases.


Topic 4: Key Points
In this section, we explored the following main points:
- 1: Epigenetic mechanisms may influence gene expression without alteration in the DNA sequence.
- 2: Main epigenetic mechanisms are DNA methylation, histones post-translational modification and alteration in the expression of microRNAs.
- 3: Epigenetic change is a regular and natural occurrence but can also be influenced by several factors including age, the environment/lifestyle, and disease state.
- 4: Different from genetic alterations, epigenetic alterations are considered reversible.
1. Epigenetic mechanisms are considered:
Reversible
Irreversible
- Answer
-
Reversible
2. DNA methylation and histone post-translational modifications play important role in the establishment of chromatin structure and in consequence in the gene expression modulation. True or False?
False
True
- Answer
-
True
3. DNA methylation may inhibit the binding of transcription factors to their recognition site, at promoter regions resulting in:
Inhibition of gene transcription
Activation of gene transcription
- Answer
-
Inhibition of gene transcription


