2.2: Cell Structure and Subcellular Compartments
- Page ID
- 308328
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- 1: Describe the basic components of cell structure and organelles.
- 2: Understand the chromatin organization into nucleus.
- 3: Understand basic mechanisms of cell signaling to respond to changes in the environment.
2.1
Structure of a cell
Cells are the smallest unit of life. A typical eukaryotic cell and it components are illustrated in the next figure.

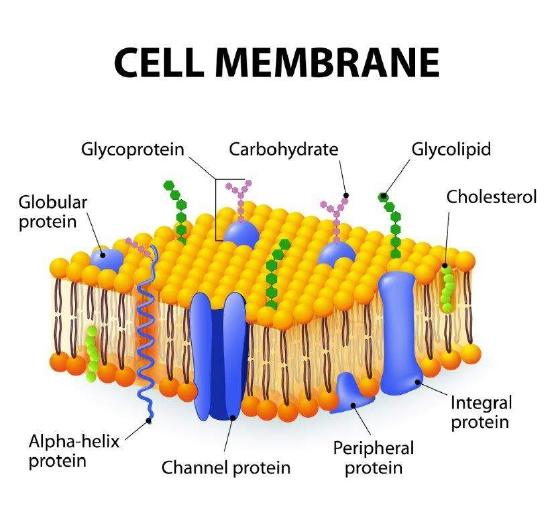 Membrane: Plasma membrane, nuclear envelope, and membranes provide separation of different environments to permit a variety of biological functions. Membranes are dynamic and fluid structures that allow selective movement of ions, energy sources, vitamins and cofactors, and waste. Membrane components include lipid, carbohydrates and protein molecules. Membrane fluidity is very important for its functions and, as a consequence, is important in disease processes, as well as, treatments.
Membrane: Plasma membrane, nuclear envelope, and membranes provide separation of different environments to permit a variety of biological functions. Membranes are dynamic and fluid structures that allow selective movement of ions, energy sources, vitamins and cofactors, and waste. Membrane components include lipid, carbohydrates and protein molecules. Membrane fluidity is very important for its functions and, as a consequence, is important in disease processes, as well as, treatments.Membranes are composed of lipids arranged in a lipid bilayer, with the hydrophilic glycerol and phosphate “head” groups of the lipid molecules forming the two outside layers and the hydrophobic “tail” groups arranged inside. Proteins are the second major part of biological membranes and makeup approximately 20%–80% of both the structural and functional components of these membranes. Many of these proteins are embedded into the membrane and stick out on both sides; these are called transmembrane proteins.
2.2
Subcellular compartments

CYTOSKELETON
is a structure that helps cells maintain their shape and internal organization, and it also provides mechanical support that enables cells to carry out essential functions like division and movement. The cytoskeleton is made up of microtubules, actin filaments, and intermediate filaments.
 ENDOPLASMIC RETICULUM (ER)
ENDOPLASMIC RETICULUM (ER)
It consists of a system of sac- and tube-like structures, which locally expand into cisterns. Its internal lumen is connected with the intermembrane space of the nuclear membrane. Part of the ER is studded on the outside with ribosomes (rough ER), which take part in protein synthesis. The other part of the ER is free of ribosomes (smooth ER). Enzymes of the smooth ER are involved in the synthesis of fatty acids. The smooth ER also plays a role in detoxification by hydroxylation reactions.
This organelle consists of stacks of flattened membrane sacs. Their main function is the further processing and sorting of proteins and their export to the final targets. In most cases, these are secreted as membrane proteins. In addition, the Golgi apparatus also produces polysaccharides.
Lysosomes are vesicles enclosed by a lipid bilayer. These organelles are filled with many enzymes for polysaccharide, lipid, protein and nucleic acid degradation. They act also on intracellular material to be removed and even contribute to the apoptosis of their own cell. Lysosomes of special cells (e.g., macrophages) destroy bacteria or viruses as a defense mechanism.
 PEROXISOMES
PEROXISOMES
Peroxisomes are surrounded by a single membrane. They are generated from components of the cytosol and do not bud from other membranes. The main task of these organelles is the performance of monooxygenase (hydroxylase) or oxidase reactions, which produce hydrogen peroxide (H2O2).
 MITOCHONDRIA
MITOCHONDRIA
In a typical eukaryotic cell, there are in the order of 2000 of these organelles, which are often of ellipsoidal shape. They have a smooth outer membrane and a highly folded inner membrane with numerous invaginations (cristae), which contain most of the membrane-bound enzymes of mitochondrial metabolism. Mitochondria are the site of respiration and ATP synthesis, but also of many other central reactions of metabolism, e.g., citrate cycle, fatty acid oxidation, glutamine formation, and part of the pathway leading to steroid hormones. Mitochondria are the only organelles which are equipped with their own (circular) DNA, RNA and ribosomes and thus can perform their own protein synthesis.
 NUCLEUS
NUCLEUS
All eukaryotic cells show the presence of a separate nucleus, which contains the major portion of the genetic material of the cell (DNA). The nuclear DNA is organized in a number of chromosomes. The nucleus is surrounded by a double membrane of lipid bilayers with integrated proteins, called the nuclear membrane (also known as the nuclear envelope). Nuclear pores span the nuclear membrane and enable the transport of proteins, rRNA etc.
Image credits
2. DataBase Center for Life Science (DBCLS) licensed under CC BY 4.0
3. Kelvin Song licensed under CC BY 3.0
4. Lumoreno licensed under CC BY-SA 3.0
5. Agateller licensed under CC BY-SA 3.0
2.3
Structure of Chromatin
Chromosomal DNA is packaged inside microscopic nuclei by its association of histones H2A, H2B, H3, and H4. These are positively-charged proteins that strongly adhere to negatively-charged DNA and form complexes called nucleosomes. (11-nm “beads on a string” structure). Each nucleosome is composed of DNA wound 1.65 times around eight histone proteins. Nucleosomes fold up to form a 30-nanometer chromatin fiber, which forms loops averaging 300 nanometers in length. The 300 nm fibers are compressed and folded to produce a 250 nm-wide fiber, which is tightly coiled into the chromatid of a chromosome. The level of structure varies depending on the cell cycle stage and, as a result, the requirement for DNA transcription or replication.
 Figure \(\PageIndex{2}\): Chromosomes are composed of DNA tightly-wound around histones - © 2010 Nature Education
Figure \(\PageIndex{2}\): Chromosomes are composed of DNA tightly-wound around histones - © 2010 Nature Education
Chromatin: The most important structure inside the nucleus is chromatin, consisting, in humans, of the 46 chromosomes. Chromatin is the combination or complex of DNA and proteins that make up the contents of the nucleus of a cell. The most abundant protein in the nucleus are histones. Histones are rich in basic amino acids (positively charged), which interact with negative charges of the DNA.
2.4
Cell Signaling
 Cell signaling consists in the ability of cells to respond to environment changes through signals received outside their borders. Cells may receive many signals simultaneously and also send out messages to other cells.
Cell signaling consists in the ability of cells to respond to environment changes through signals received outside their borders. Cells may receive many signals simultaneously and also send out messages to other cells.
Cells have proteins called receptors, that are generally transmembrane proteins, which bind to signaling molecules outside the cell and subsequently transmit the signal through a sequence of molecular switches to internal signaling pathways and initiate a physiological response. Different receptors are specific for different molecules. In fact, there are hundreds of receptor types found in cells, and varying cell types have different populations of receptors.
Figure \(\PageIndex{3}\): Main types of transmembrane receptors
Examples of receptors membrane:
- G-protein-coupled receptors
- Ion channel receptors
- Enzyme-linked receptors
Receptor may be located in the cellular membrane (important to receive extracellular signals) but also may be present inside the cell or inside the nucleus.
Topic 2: Key Points
In this section, we explored the following main points:
- 1: The characteristics of the cell membrane, such as lipid bilayer and transmembrane proteins are very important for the cell function.
- 2: The chromatin is the combination of complex DNA and proteins that make up the contents of the nucleus of a cell.
- 3: Cells have proteins called receptors that are important to receive extracellular signals and initiate signaling pathways in the cell.
1. Histones are proteins present in the nucleus of the cells with the function of:
interacting with the DNA to form nucleosomes.
regulating the fluidity of cell membranes.
- Answer
-
interacting with the DNA to form nucleosomes.
2. Membranes are composed of lipids arranged in a ...
Check all that apply.
- lipid monolayer, with...
- lipid bilayer, with...
- ...the hydrophilic glycerol and phosphate “head” groups of the lipid molecules forming the two outside layers and the hydrophobic “tail” groups arranged inside.
- ...the hydrophilic and hydrophobic “tail” groups arranged inside.
- Answer
-
lipid bilayer, with...the hydrophilic glycerol and phosphate “head” groups of the lipid molecules forming the two outside layers and the hydrophobic “tail” groups arranged inside.


