Applications
- Page ID
- 272754
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Purify a protein of interest
A major problem in the field of molecular biology involves the expression and purification of a protein of interest, for example the human adenovirus serotype, Ad5. This protein has been commonly used as a vector for gene therapy, due to its efficient gene delivery and application in a diverse range of cells. However, limitations have been observed in acquiring the necessary expression yields and in purifying the collected proteins. These problems have been addressed by Chen et al. through the construction of a fusion of Ad5 with an N-terminal 6-Histidine (His) tag, expressed in E. Coli. This fusion shows high expression levels, within 5 h of induction. A simple one step metal affinity purification system, utilizing the His tag which binds to Ni-NTA, was used to isolate the Ad5, with greater than 95% purity. The isolated protein was evaluated via a receptor-binding assay, in Hela cells.
Reference: Juyun Yang, Yanzhong Zhao, Ting Liu, Yuxiang Chen, and Shuyi Yu, High-Level Expression, One-Step Purification of Soluble Ad5-Knob Protein and Its Activity Assay Cancer Biotherapy and Radiopharmaceuticals, 2006, 21, 269-275
Purify protein of interest based on its inherent ligand-binding specificity
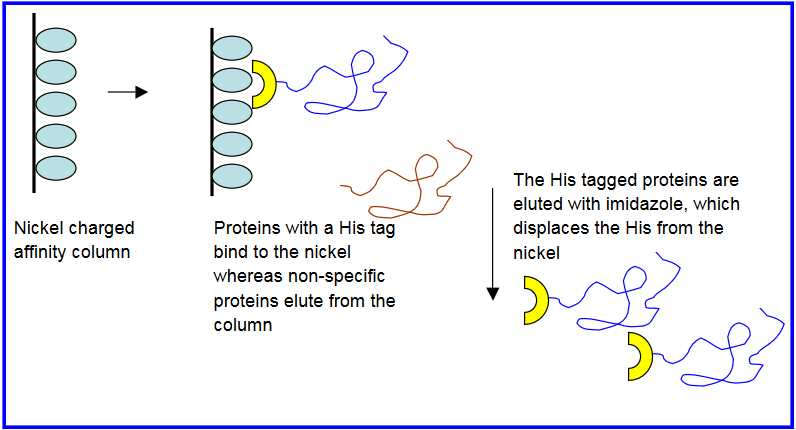
There are a few proteins that have natural affinity to bind metal ions. These proteins are typically metal chaperone proteins. This inherent metal ion binding has been utilized as a way to purify these proteins using metal immobilized affinity columns. For example, a red fluorescent protein, DsRed, has a binding affinity for copper ions. On the basis of this a purification method for DsRed was developed using copper immobilized column. A metal chelating ligand attached to beads was used in this study to immobilize copper ions on beads. The crude protein was passed through the copper immobilized beads. DsRed bound to the beads, whereas other interfering proteins did not bind to the beads and hence were removed. DsRed was eluted using a competitive ligand imidazole that binds to copper ions. This is a simpler approach of protein purification based on their inherent metal ions affinity.
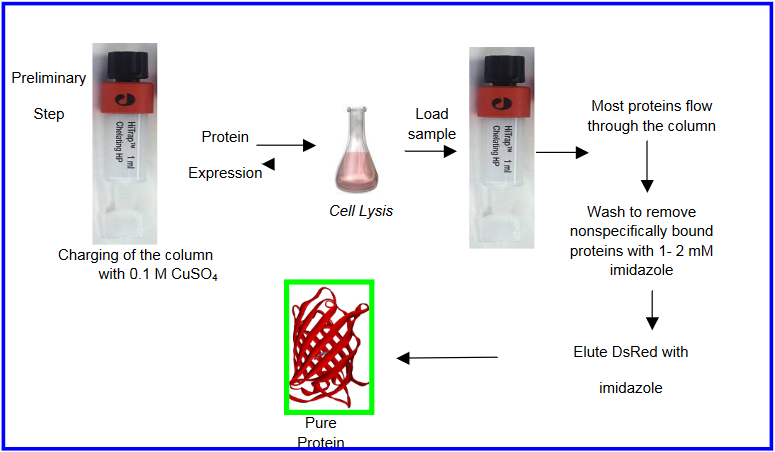
Reference: Y. Rahimi, S. Shrestha and S. K. Deo, Metal Affinity-Based Purification of a Red Fluorescent Protein Chromatographia, 2007, 65, 2007, 429-433
Affinity-based chiral separations
The stereochemistry of a compound often determines its biological activity, for this reason chiral separation of compounds is an important aspect of the drug discovery process. Wainer and co-workers developed a chiral affinity stationary phase, a-chymotrypsin, with which they were able to resolve D,D- and L,L-dipeptides and D,D-/L,L- and L,D-/D,L-dipeptides. The results of these studies have, also, suggested the possibility of multiple binding site interactions between the target dipeptide and the a-chymotrypsin.
Study of drug interactions
The quantitative characterization of interactions between targets and ligands is an integral part of the drug discovery process. These interactions are often defined by the association constant of a drug-protein interaction. Kim et al. utilized affinity chromatography to define the association constants, for a number of HSA-binding drugs, based upon their retention factors. They prepared an immobilized HSA column, by passing known concentrations of the drugs through the column, saturation plots were generated which were used to calculate retention factors and association constants. This system offers advantages over the more common methods of evaluating association constants; such as fast response times, ease of automation, and additionally the ability to distinguish chiral compounds simultaneously.

Reference: Hee Seung Kim and Irving W. Wainer, Rapid Analysis of the Interactions between Drugs and Human Serum Albumin (HSA) Using High-Performance Affinity Chromatography (HPAC) Journal of Chromatography B, 2008, 870, 22-26.


