15: IMF of BioMolecules
- Page ID
- 142575
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Structure of Biomolecules: Examining the Role of Intermolecular Forces
Four Categories of Biological Molecules
-
Sugars
-
Lipids
-
Proteins
-
DNA&RNA
Polymers and Supramolecular Structures
Polymer: A polymer is a large molecule composed of many repeated subunits. the repeating subunits (monomers) are linked through covalent bonds. the most common macromolecules in biological systems are polymers
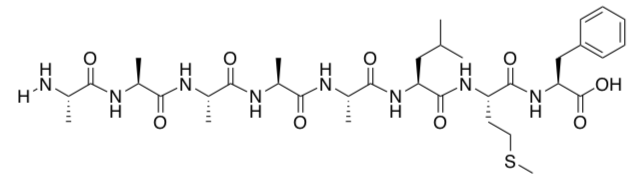
Proteins are large molecules made of amino acids which link covalently to form a polymer.
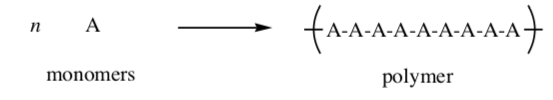
- Identify the repeating units in this polypeptide.
Supramolecular Assembly: A supramolecular assembly is a complex of molecules held together by intermolecular forces. These complexes are common in biological systems. Micelles, membranes, liposomes and the DNA double helix are all examples of supramolecular assemblies.
Here is a cartoon of an enzyme:substrate supramolecular complex.

-
Considering how these structures are held together, which would be stronger – polymers OR supramolecular assemblies? Circle one.
-
Explain why biological systems employ both polymers and supramolecular structures. For example, proteins are polymers but an enzyme:substrate complex is a supramolecular assembly.
I. Sugars -> Polysaccharides
Carbohydrates, commonly called sugars, are polyhydroxy straight chain aldehydes or ketones with numerous chiral centers (as seen in previous sections). These sugars can cyclize to form rings also.
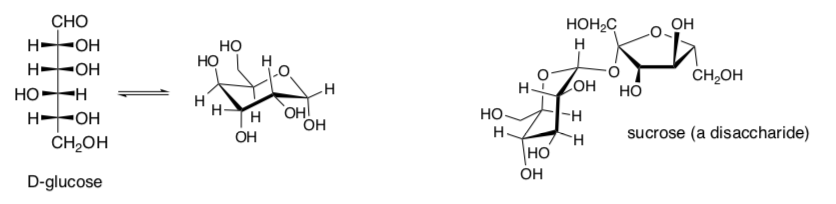
-
Based on the structures above, would you predict most simple sugars to be soluble in water?
-
Sugars provide energy for cells throughout the body. Explain why solubility is important.
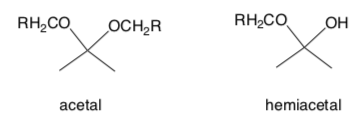
A complex carbohydrate, a polysaccharide, is several sugars joined by an acetal linkage.
- Circle or highlight the acetal functional group in the cyclic structures of D- glucose and sucrose above.
Many toxic compounds or pharmaceuticals are glycosylated (a glucose is added) during metabolism in the body. Here is an example of a barbiturate after glycosylation.
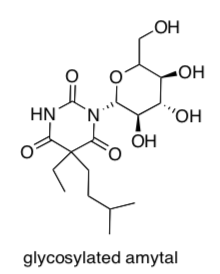
- Circle the sugar that was added to the molecule.
- Does this glycosylation [ increase or decrease] the polarity of the molecule?
- Why does glucose get added before the compound can be excreted in the urine?
Polysaccharides: Starch
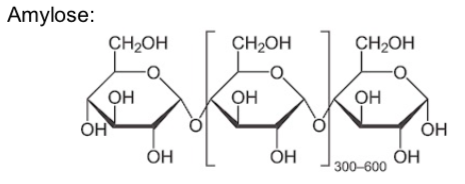
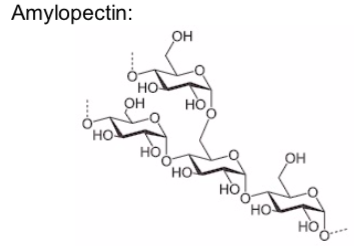
The polysaccharide known as starch is the major food reserve found in plants. Plant starch is made of the polymers, amylose and amylopectin.
Amylose is made up of (1→4) linked glucose molecules. The number of repeated glucose subunits (n) is usually in the range of 300 to 3000, but can be many thousands.
Amylopectin is a polysaccharide and highly branched polymer of glucose found in plants. It is one of the two components of starch, the other being amylose.
- Carbohydrates are an example of a [ polymer or a supramolecular assembly ] ? Circle one.
Amylose is important in plant energy storage. It is less readily digested than amylopectin.
-
Amylopectin is more soluble in water than amylose. Propose a reason why.
-
Storage space is always a limiting factor (whether its for shoes or starches). Based on the shape of the two starch components, which would be preferred for storage? (ie which takes up less space?)
amylose or amylopectin
Carbohydrate Application Problems:
-
Raffinoseisatrisaccharidecomposedofgalactose,fructose,andglucose.Itcan be found in beans, cabbage, other vegetables, and whole grains. Raffinose can be digested to D-galactose and sucrose by the enzyme α-galactosidase (α-GAL), an enzyme not found in the human digestive tract.
In the lower intestine, they are fermented by gas-producing bacteria which do possess the α-GAL enzyme and make carbon dioxide and/or methane.
-
What is the active ingredient in ‘Beano’, a dietary supplement intended to reduce flatulence?
-
-
Wheat kernels are the seeds of the plant. Inside the kernel is a large quantity of starch that is the stored food source for the developing plant. Upon germination the seedling secretes enzymes which breakdown the starch into its simple sugar units thus making them available to the developing plant. In modern wheat production, the wheat is harvested before these enzymes have been secreted.
Most yeast (needed for fermentation) also lack the enzyme, α-amylase, that is responsible for the breakdown of the starch molecule. α-Amylase is very similar to α-galactosidase (α-GAL).
Biochemist Joseph Owades revolutionized the production of beer by developing a modification of the fermentation process that converted regular beer into “light” beer.
-
What was the modification?
-
How does light beer differ from regular beer?
-
If you are home-brewing beer and don’t have access to Owades’ commercial modification, what could you use instead?
-
-
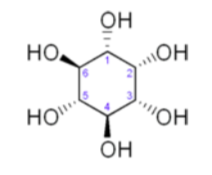
Myo-Inositolactsasagrowthfactorinbothanimalsandmicroorganisms.
-
Draw the most stable chair conformation of this compound.
-
-
Galactose is a type of sugar molecule, shown below:
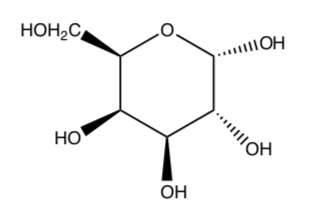
-
Circle all the chiral centers in the galactose molecule.
-
How many possible stereoisomers of galactose are there?
-
Draw galactose in its most stable chair conformation.
-
What is the relationship between a galactose molecule and a glucose molecule (structure shown below)?
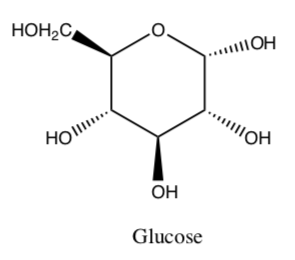
-
II. Lipids -> Micelles and Lipid Bilayers
Lipids
Lipids are a class of biological compounds that are soluble in organic solvents. There are a variety of structures that fulfill this requirement.
- To be soluble in organic solvents, lipids all have some [ hydrophobic or hydrophilic ] components to their structure. Circle one.
Several examples of lipids are shown below.
- Categorize each lipid as hydrophobic (nonpolar) or amphiphilic (non-polar regions and polar regions).
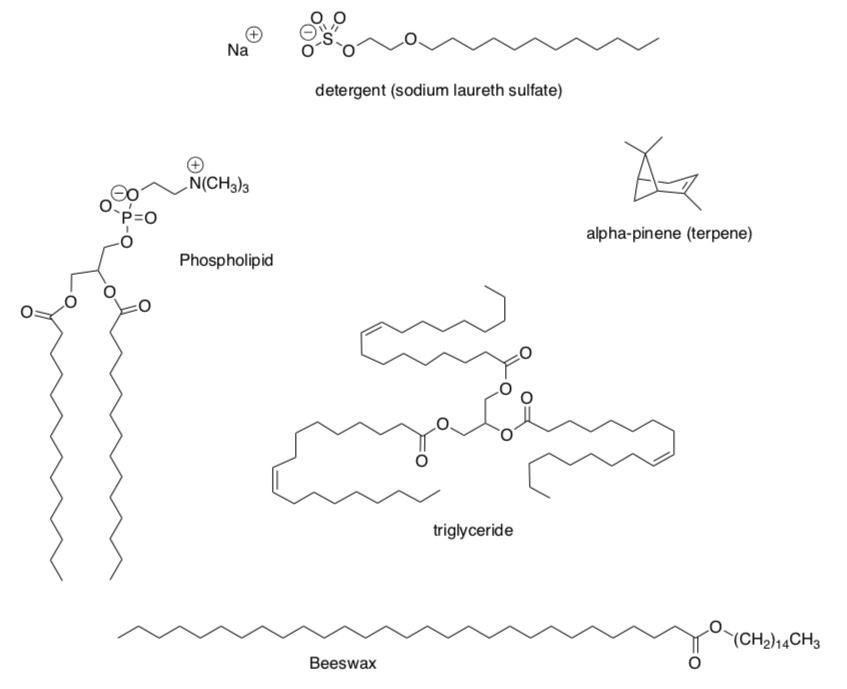
Lipid Aggregates: Micelles and Membrane Bilayers
The amphiphilic lipids can be drawn in “cartoon” form as a circle representing the polar end and a squiggly line representing the nonpolar end.
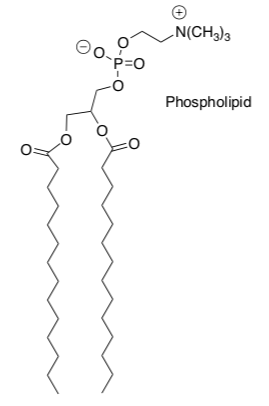
Phospholipids (shown below) are typical amphiphilic lipids.
-
Circle the polar end.
-
Draw a squiggly line beside the hydrophobic region.
When amphiphilic lipids are mixed with water, they form aggregates such as lipid bilayers or micelles.
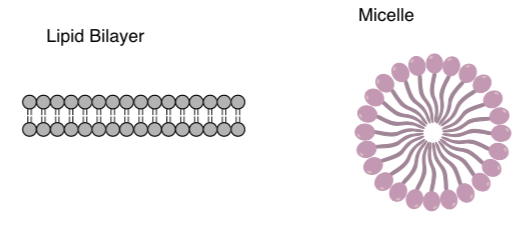
- Add several waters to these two pictures.
Identify the IMF in Lipid Aggregates:
There are several IMFs involved in the phospholipid bilayer or micelle.
-
For each of the following interactions, circle the predominant intermolecular interaction.
-
The head: head interaction is:
dipole-dipole London Dispersion ion-dipole
hydrogen bonding ion-ion
-
The tail: tail interaction is:
dipole-dipole London Dispersion ion-dipole
hydrogen bonding ion-ion
-
The water: head of the phospholipid interaction is:
dipole-dipole London Dispersion ion-dipole
hydrogen bonding ion-ion
-
The water: tails of phospholipid is
attractive repulsive
-
-
Lipid bilayers and micelles are examples of [ polymer or a supramolecular assembly ] ? Circle one.
Lipid Aggregates: Micelles and Membrane Bilayers
-
What role does surface tension play in the formation of micelles?
-
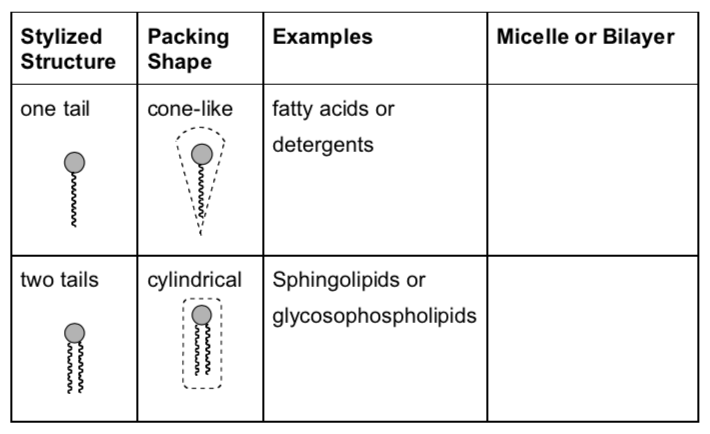
Complete the data table below to predict what type of lipids will form micelles and which lipids will be more likely to form bilayers.
Properties of Cell Membranes: Fluidity
The cell membrane is a lipid bilayer that separates the interior of all cells from the outside environment.
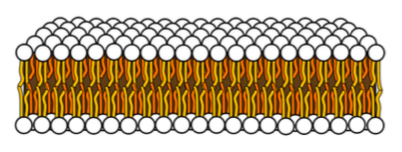
The cell membrane is partially fluid – able to move.
-
Why does the cell membrane need to be fluid in most organisms?
-
How does the structure provide fluidity (i.e. consider how the structure is held together)?
The cell membrane is not as fluid as a liquid.
-
Why does the cell need some rigidity?
Cell membranes are self-sealing. If they are disrupted, they quickly and spontaneously reseal.
- Explain what is happening using your understanding of intermolecular forces.
Properties of Cell Membranes: Permeability
The cell membrane controls the movement of substances in and out of cells, thus facilitating the transport of materials needed for survival.
-
Explain the importance of regulating the type and amount of substances entering the cells.
Some mechanisms to enable substances to move across the cell membrane:
-
Passive diffusion and osmosis.
Some substances such as carbon dioxide (CO2) and oxygen (O2), can move across the cell membrane by diffusion.
- Why can CO2 and O2 move through the lipid bilayer? Use relevant IMF in your explanation.
Some small molecules can also move through cell membrane.
-
What would you predict about the polarity of the molecules that can diffuse the lipid bilayer?
small non-polar large nonpolar polar polar charged ions gases.
-
Explain your choice.
-
Would glucose passively diffuse through the bilayer?
-
Transmembrane channels, transporters and pumps.
These ‘holes’ in the cell membrane regulate the passage of all substances that can’t simply diffuse through the cell membrane.
-
What would you predict about the polarity of the molecules that need a special channel or transport mechanism? Circle all that apply.
small non-polar large nonpolar polar polar charged ions gases.
-
Explain your choice.
-
Cholesterol in Cell Membranes
The cholesterol molecule inserts itself in the membrane because it also has some amphiphilic character.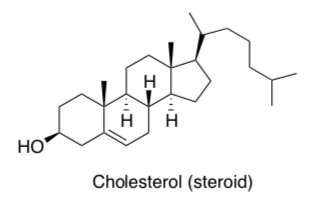
-
Circle the polar end.
-
Put a box around the non-polar end.
The cholesterol molecule inserts itself in the membrane with the same orientation as the phospholipid molecules.
-
Add a cartoon of the cholesterol to the lipid bilayer below.
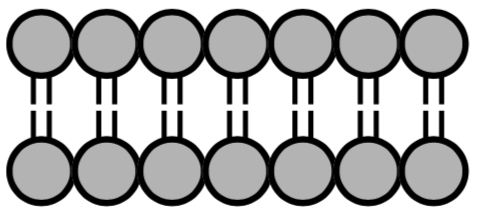
There are several IMFs involved in the insertion of the cholesterol into the phopholipid bilayer of a cell membrane or liposome.
-
For each of the following interactions, circle the predominant IMF.
-
The head:head interaction is:
dipole-dipole London Dispersion ion-dipole hydrogen bonding ion-ion
-
The tail: tail interaction is:
dipole-dipole London Dispersion ion-dipole hydrogen bonding ion-ion
-
The water: head of the cholesterol interaction is:
dipole-dipole London Dispersion ion-dipole hydrogen bonding ion-ion
-
The water: tails of cholesterol is
attractive repulsive
-
Function of cholesterol in the Cell Membrane
Cholesterol molecules have several functions in the membrane.
- Draw pictures and use words to explain the three functions listed below:
-
Cholesterol decreases membrane fluidity at high temperatures. Explain how the addition of cholesterol to the lipid bilayer would decrease membrane fluidity at high temperatures.
-
Cholesterol prevents crystallization of hydrocarbons in the membrane at low temperature. Explain how the addition of cholesterol to the lipid bilayer prevents crystallization of the alkyl chains.
-
Sterols decreases the permeability of the cell membrane to small water- soluble molecules. Explain how the addition of cholesterol to the lipid bilayer would decrease the permeability of the bilayer to water and water- soluble compounds.
-
Lipid Application Questions:
-
Molecular Solids/IFats
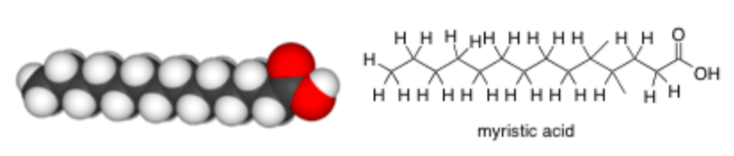
Saturated fats have no double bonds and are sometimes associated with cardiovascular disease. Examples of foods containing a high proportion of saturated fat include dairy products (especially cream and cheese but also butter and ghee); animal fats such as suet, tallow, lard and fatty meat; coconut oil, cottonseed oil and palm kernel oil.Myristic Acid (shown below) is an example of a saturated fat from coconuts.
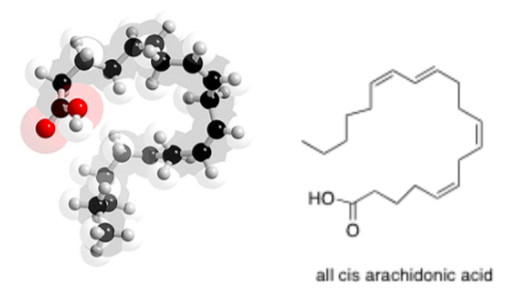
Unsaturated fats have at least one double bond. Examples of foods containing a high proportion of unsaturated fats include avocados, olives, and canola oil.
Arachidonic Acid (shown below) is an unsaturated fat from peanut oil.
-
Which is more likely to pack together tightly to form a molecular solid?
Saturated Fats Unsaturated Fats
-
Explain your answer.
-
-
Lipids, like carbohydrates, are often used as an energy source in the body. However, by definition, lipids are not soluble in water. Suggest a method for distribution throughout the body.
-
Snake venom contains the enzyme phospholipase A2. The action of phospholipase A2 cleaves the fatty acid tail from the phosphate ester head of the phospholipid structure. Discuss how this will affect the cell membrane.
-
Some fats are optically active while others are optically inactive.
-
Draw one type of lipids that are achiral.
-
Draw a chiral lipid.
-
III. Amino Acids → Proteins
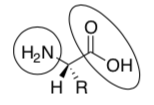
Passport: Amino acids all have the same generic structure:
-
What is the functional group in the circle?
-
What is the functional group in the oval?
-
Put the chiral center in a box.
-
Redraw the amino acid above where the R group is a methyl (CH3). This is alanine.
-
Identify whether alanine is R or S.
There are 20 different naturally occurring amino acids, each with a different R group
(see Table in the front of this workbook)
-
Identify an example of an amino acid side chains that is nonpolar.
-
Identify an example of a positively charged side chains.
-
Identify an example of a negatively charged side chains.
-
Identify an example of an amino acid side chains that is polar uncharged.
-
Which of these types of side chains are hydrophilic (water loving):
nonpolar positively charged negatively charged polar uncharged
-
Which of these types of side chains are hydrophobic (water hating):
nonpolar positively charged negatively charged polar uncharged
Protein Structure
There are four distinct levels of protein structure.
-
The primary structure is the sequence of different amino acids in a protein.
-
Secondary structure refers to highly regular local sub-structures. Two main types of secondary structure, the alpha helix and the beta strand.
-
Tertiary structure is the three-dimensional structure structure of a protein molecule.
-
Quaternary structure is a larger assembly of several protein molecules.
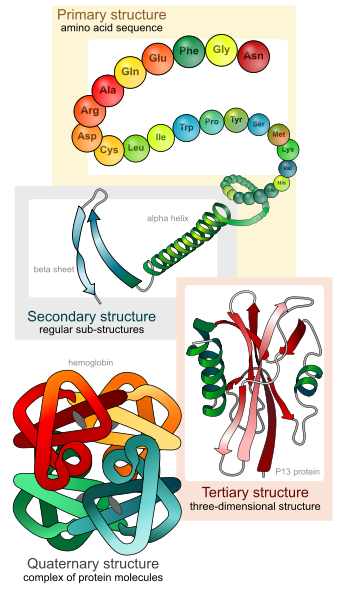
From: Wikipedia, http://en.wikipedia.org/wiki/File:Ma..._levels_en.svg
1. Primary Structure:
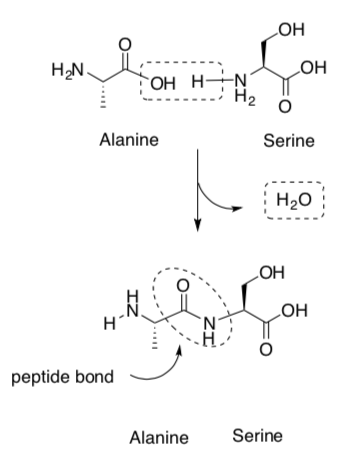
Proteins are large molecules made of amino acids which link covalently to form a polymer:
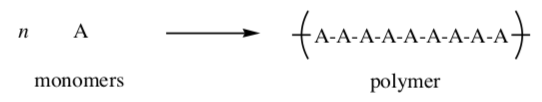
The primary structure refers to linear sequence of the amino acid chain. The primary structure is held together by covalent bonds (the peptide bond).
-
What is the functional group formed (peptide bond)?
-
What is the side product formed?
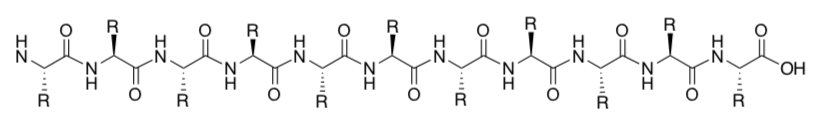
- Highlight the backbone of the protein (excluding the side chains).
-
Is the main backbone ( polar or nonpolar)? Circle one.
-
Primary protein structure is an example of a [ polymer or a supramolecular assembly ] ? Circle one.
2a. Secondary Structures – Beta Sheets
Proteins fold into one or more specific spatial conformations – secondary structure. IMF holds secondary structures into place between the backbone functional groups.
Beta sheets consist of beta strands (shown below) connected laterally by IMF, forming a generally twisted, pleated sheet.
-
What is the IMF holding these two strands together?
-
Clearly indicate on the above picture which atoms are involved in these IMFs.
Beta sheets can be parallel (two strands running in the same direction) or antiparallel (opposite directions.
-
Decide which of the pictures below are parallel and which is antiparallel.
2b. Secondary Structures: Alpha Helices
The alpha helix (α-helix) is another example of secondary structure of proteins.
-
The alpha helix is a spiral conformation in which every backbone of the protein completes a full spiral every ___________residues.
Again, intermolecular forces from the backbone of the protein hold this structural motif in place.
-
What is the IMF holding this strand in a coil?
-
Clearly indicate on the picture above which atoms are involved in these IMFs.
2c. Secondary Structures: Alpha Helices
A helical wheel plot is a two-dimensional representation of the residues of an alpha helix where you are looking down the axis.
This is a helical wheel for the sequence:
abcdeABCDE123456789
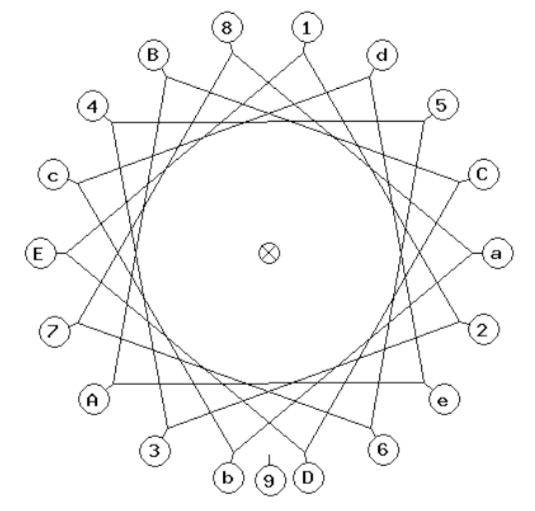
Here is a real sequence that forms an alpha helix:
Leu- Glu-Glu-Val-Phe-Ser-Gln-Leu-Cys-Thr-His-Val-Glu-Thr-Leu-Lys
For the amino acids in this sequence:
-
Create your helical wheel by plotting the position of each amino acid on the helical wheel plot above including the circles and boxes. Start with the leucine at a. Continue to place the other amino acids around the wheel.
-
If possible, also create a handheld model using pipecleaners and a Styrofoam tube.
-
Identify the hydrophilic residues by circling them.
-
Identify the hydrophobic residues by boxing them.
-
Which face of the alpha helix is most likely to face a water solution?
-
Which face of the alpha helix is most likely to face the interior of the protein?
3. Tertiary Structures
The packing of protein secondary structure elements into compact globular units forms tertiary structure of a protein. Proteins are packed such that very little water is found inside the folded protein. The folding is driven by the non-specifichydrophobic interactions (the burial of hydrophobic residues from water).
-
[Hydrophobic or hydrophilic ] amino acids are more likely to be found on the surface of the protein. Circle one.
-
[Hydrophobic or hydrophilic ] amino acids will be most likely to be found buried in the folded protein. Circle one.
-
If a Val side chain is bound buried in the protein (not exposed to water), which other amino acid side chains might be around it?
4. Quaternary Structures
Quaternary structure is the 3D structure of a multi-subunit protein and how the subunits fit together.
-
If this is a monomer, draw a cartoon of a tetramer.

-
Subunits are held together including salt bridges and hydrogen bonding. Identify these IMF attractions on the picture.
-
Quaternary protein structure is an example of a [ polymer or a supramolecular assembly ] ? Circle one.
Protein Folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil.
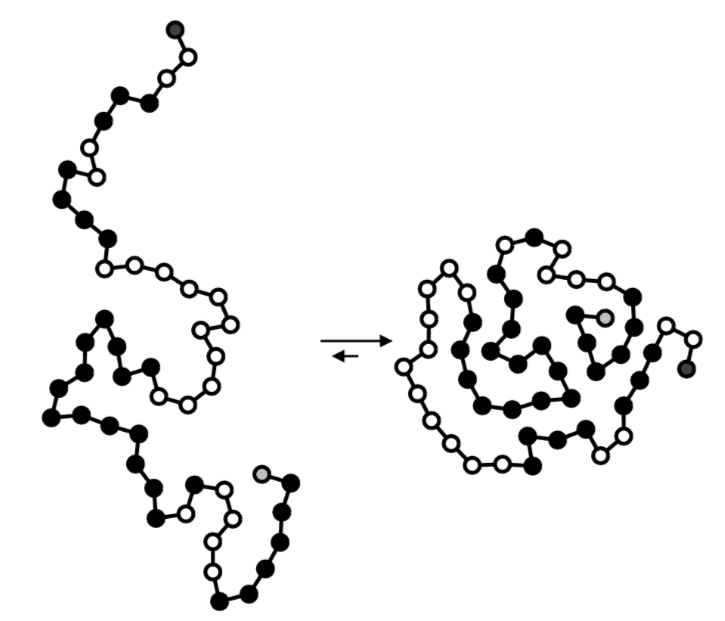
From Wikipedia, http://upload.wikimedia.org/wikipedi..._schematic.png
- If the black residues are hydrophobic, describe one main driving force for protein folding.
Protein Denaturation
Denaturation is a process in which proteins lose their secondary or tertiary structure that is present in their folded state.
-
Draw a cartoon for the denaturation process.
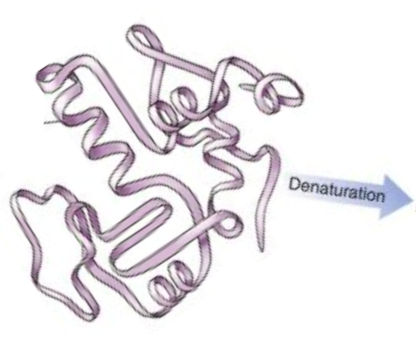
-
On a molecular level, describe what happens when a protein is denatured.
-
Suggest 2-3 different things that might cause denaturation.
-
If proteins in living cells are denatured, what might happen to the cell?
Protein Practice Problems
-
Draw all possible of stereoisomers of threonine and assign R, S nomenclature to each.
-
Draw the structure of the following peptide:
Val-Glu-Pro-Ala-Cys
-
The \(\alpha\) -helical segments of proteins stop whenever a proline is encountered in the chain. Suggest a reason why proline isn't found in an \(\alpha\)-helix.
-
Pro is the amino acid most commonly found in \(\beta\)-turns. Discuss the reasons for this observation.
-
Foldit!
Foldit is a revolutionary new computer game enabling you to contribute to important scientific research.
https://fold.it/portal/info/about
Dartnell, Lewis New Scientist. 11/8/2008, Vol. 199 (2681), p36-39.
For this assignment:
-
Read about the protein folding game at the website and/or the New Scientist article.
-
Attempt to fold a protein. Check to see if your instructor has set up a private group! https://fold.it/portal/node/996074
Turn in:
-
A summary (in your own words) answering the following questions:
-
Why are scientists interested in protein folding?
-
Why make it a computer game rather than just have a computer solve the structure?
-
What protein did you attempt to fold?
-
Why was your protein important?
-
Provide a screen shot of your attempt. (Or if your instructor has made a private group, they will be able to follow your attempts).
-
-
Receptor Theory
The active site is the 3-D groove or pocket of an enzyme or receptor where molecules bind and undergo a chemical reaction or trigger a response.
In order to interact with the active site, the substrate:
a) has to be held into the active site with intermolecular forces with the amino acid side chains.
b) must “fit” into the active site. (size and geometry and orientation)
a. Binding: Electrostatic Forces
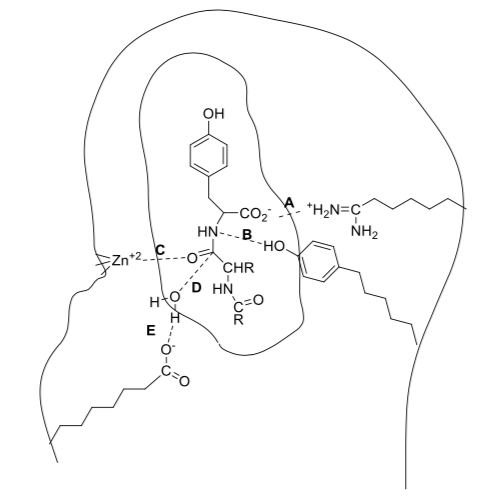
Substrates bind to receptors and enzymes either by covalent or non-covalent interactions.
Below is a cartoon of a part of a peptide molecule (inner circle) interacting with an enzyme. The dashed lines show intermolecular forces holding the substrate to the enzyme.
- Label one example of an intermolecular force shown.
b. Binding: Fit, Size and Geometry
Receptors and enzymes have 3-dimensional shape so the structure of the substrate must fit.
- Based on size and geometry, which substrate will provide the tightest fit
c. Binding: Stereochemistry
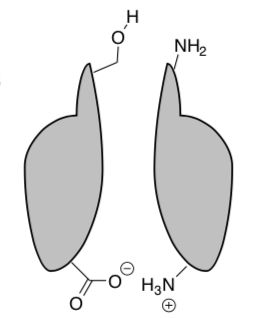
In addition, stereochemistry makes a big difference. Below is a cartoon of two enantiomers and how they interact with a receptor.
When structure A binds, it takes full advantage of all the possible interactions (much like a hand in a glove or a key in a lock – it fits perfectly). Its enantiomer, structure B, is not able to take advantage of all the interactions.

-
Is Structure A or structure B bound more tightly? Circle one.
-
Draw the Newman projection of Structure B rotated 60, 120 and 180 degrees.
-
Is any conformation of Structure B able to take full advantage of all the possible binding pockets?
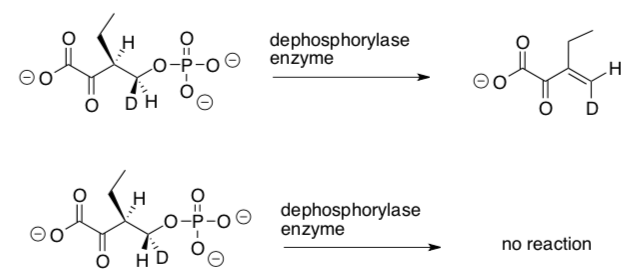
Only one isomer of the deuterated compound below is a substrate for a dephosphorylase that gives anti elimination to the Z-isomer (also shown below).
-
Label the stereocenters with R or S on these two isomers.
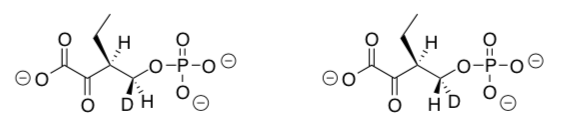
-
How are these isomers related to each other?
enantiomers diastereomers conformers
-
Using receptor theory, explain why only one of these isomers reacts with the enzyme to form a product.
Protein Application Problems
-
Nicotinic receptor binding
The more interactions that a ligand can have with a receptor or enzyme the stronger they will bind to one another.
Acetylcholine (structure shown below) interacts with a recognition site on the a subunit of the nicotinic receptor.
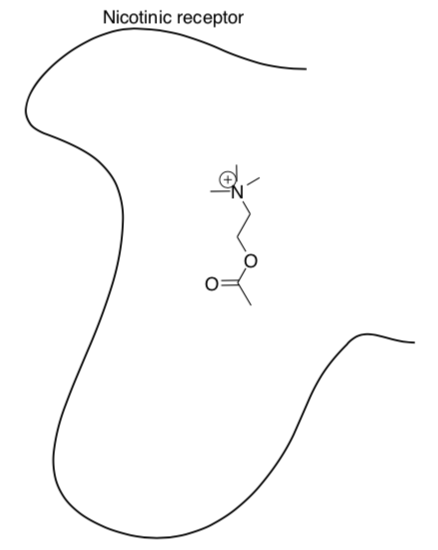
- Draw in some amino acid residues on the nicotinic residues that can form
-
ion-ion interactions
-
hydrogen bonding
-
dipole-dipole interactions
-
-
Show the interactions with acetylcholine.
- Draw in some amino acid residues on the nicotinic residues that can form
-
Stereochemistry of Drug Design.
Read the following article about stereochemistry of a drug: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC353039/
-
Draw a cartoon that represents how one enantiomer of a chiral drug will bind more tightly to an enzyme.
-
Provide an example of a current medication that has only one enantiomer that is active.
-
Currently, there is no regulatory mandate in the United States or Europe to develop new drugs exclusively as single enantiomers – the pharmaceutical company gets to choose whether to pursue a racemic or pure enantiomer for a new drug. Make an argument whether or not the FDA should require enantiomerically pure drugs for market.
-
-
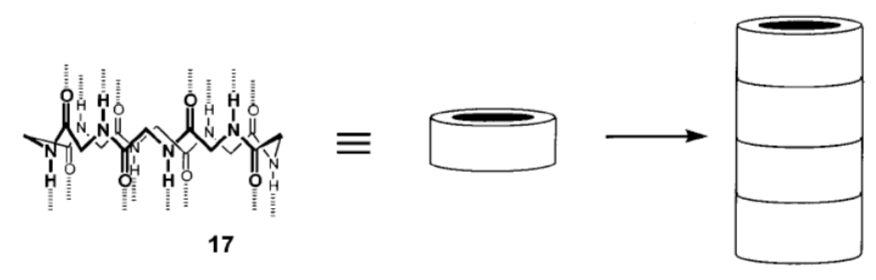
Nanotubes from Cyclic Peptides as Membrane Ion Channels
The cyclic peptides were shown to self-assemble in cell membranes to form trans-membrane size-selective ion channels and transport biologically relevant molecules, such as glucose across the lipid bilayer. Biophys J. 2003 October; 85(4): 2287–2298; Chem. Soc. Rev., 2012, 41, 6023–6041; J. Am. Chem. Soc. 1998, 120, 4417-4424
Cyclic peptide self assembly
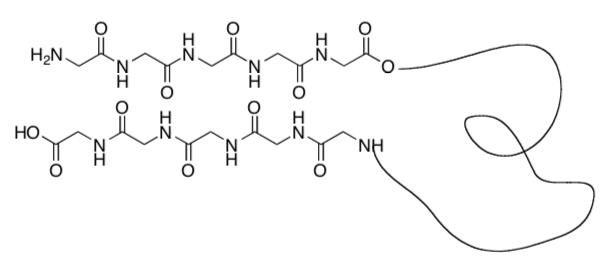
N-methylation, replacing some of the H’s on the nitrogen with CH3, limits the self- assembly process to two subunits (only forms a dimer, see image below).
-
What is holding the stackable cyclic peptides together (see image below)?
-
Indicate this in the picture below using dashed lines or highlighter.
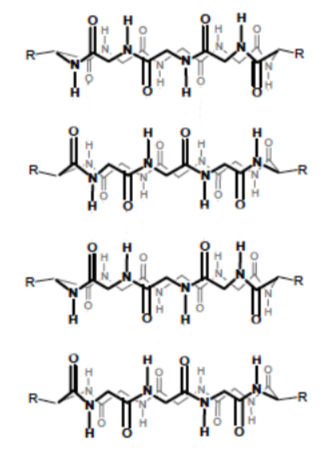
-
Explain why only two subunits can bind together after N-methylation.
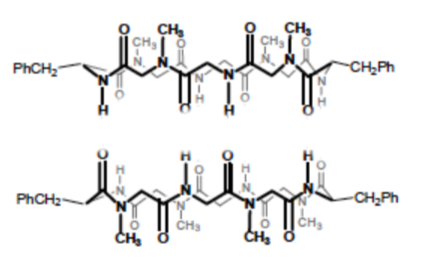
-
-
Nanotubes (cont.): Cyclic peptide design
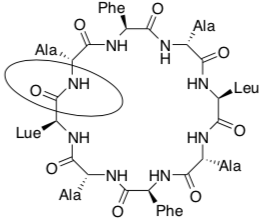
-
Name the circled functional group in the cyclic peptide shown above.
-
How many chiral centers are there on the cyclic peptide above?
-
How many stereoisomers are possible for this cyclic peptide?
-
In the box below, using wedges and dashes draw two alanine (Ala) side chains, both as the R stereoisomer, on the peptide backbone below.
-
In the circle below, using wedges and dashes draw two alanine (Ala) side chains, one as the R and one as the S stereoisomer, on the peptide backbone below.
-
The goal is to make make hollow tubes (nanotubes). Using your pictures above explain why they need to switch between R and S amino acids as they go around the cyclic peptide.
-
Why is each cyclic peptide made up of an even number of amino acids?
-
-
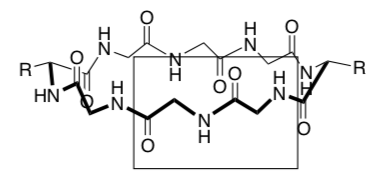
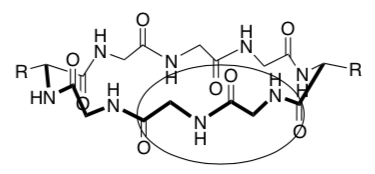
Nanotubes (cont.): Formation of channels through cell membrane
Supramolecular stacked cyclic peptides are used to create ion and transport channels (tubes) through the cell membrane.
Amino acids that are commonly used to make the cyclic peptide are phenylalanine (Phe), alanine (Ala), leucine (Leu).
-
These amino acids have side chains that are: (circle correct answer)
positively charged negatively charged nonpolar polar
-
Draw two of these amino acids coming off the cylinder below. Clearly show if the amino acid side chain is interacting with the head or the tail of the cell membrane.
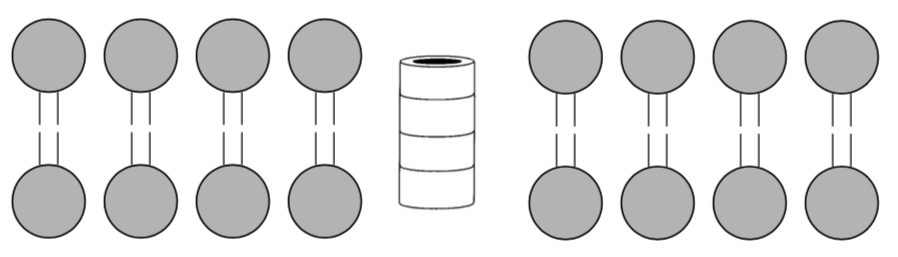
-
What type of intermolecular forces are holding the cylinders in the cell membrane?
-
-
Nanotubes (cont.): Transport of ions and glucose through the cell membrane
When a different number of amino acids are used to make the cyclic peptide the pore size (internal diameter) changes.
Number of Amino Acids Pore Size ( What fits in the channel 4 4 8 8 10 13 Current studies on these self-assembling cyclic peptide channels in cell membranes involve drug delivery and antibacterial studies.
-
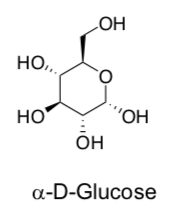
Draw glucose in its most stable chair conformation.
-
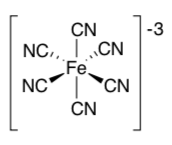
What is the geometry of Fe in Fe(CN)63- ?
-
Each of these three choices fits well into one of the channels listed above. Add these to the table.
K+ Fe(CN)63- glucose
-
Compare the size of Na+ and K+. If you wanted to design a channel selective for Na+ how many amino acids would you use?
-
-
Exploring Biological Macromolecule Interactions with Ions/Small Molecules
The biological function of macromolecules such as DNA and proteins invariably involve the binding of other molecules to them. Biochemists call these other molecules or ions ligands. The binding of ligands to proteins and nucleic acids occur through the familiar intermolecular forces you have already studied. You can visualize these interactions by viewing the 3D structure of the macromolecule:ligand complex, whose x-ray crystal coordinates are deposited in the Protein Data Bank in the form of pdb files. Do a Google search on pdb and follow the links to the RCSB Protein Data Bank. Once there, type into the PDB ID or Text search bars on the top menu the PDB # for one of the complexes shown in the tables below. Use will use a program called Ligand Explore to visualize the bound ligand..
Launch Ligand Explorer: Scroll down to the Ligand Chemical Component section in the middle of the displayed page for that complex. Select Ligand Explorer in the row with the ligand name. An example is shown below.

You may be prompted to allow pop-ups form the site. If so, allow it. You may have to reselect Ligand Explorer again to start the program. Keep giving permissions and following prompts until Ligand Explorer is open. Once launched, select the Help tab and instructions will open in a browser. Use the mouse to help find the best view of the interactions.
Assignment: Each table will select a unique PDB structure and present a one page PowerPoint in class in which you show screen shots (see below) with clear captions depicting the IMFs involved in ligand binding to your protein.
Use Ligand Explorer: on the left hand side of the Ligand Explorer window -
The oxygen and nitrogen atoms in PDB files are colored coded in most display programs as red and blue, respectively. H bonds are shown between a red O and a blue N atom, which are both electronegative and slightly negative. H atoms are not displayed in structure files obtained using X-ray crystallography. Assume that there is a H atom on one of the atoms engaged in a hydrogen bond.
Capturing cropped screen shots: PC: Select the bottom left Windows icon and type in Snipping Tool in Search Programs. Follow prompts. Mac: Select Command-Shift-4. A cross-hair cursor will appear. Click and drag to select the area. When you release the mouse button, the screen shot will be saved as a PNG file on your desktop.
A. Macromolecule: Medicinal Drug interactions
Class Macromolecule Drug Description PDB # antibiotics D-alanyl-D-alanine carboxypeptidase penicillin inhibits the enzyme that builds bacterial cell walls 1pwc antiviral HIV protease saquinavir inhibits an enzyme that is needed for HIV maturation 1hxb antiviral neuramidase tamiflu Tamiflu is an anti-influenza medicine 2hu4 anticancer DNA bleomycin binds to DNA in actively growing cells, often cleaving the DNA chain 1mxk anticancer Tubulin taxol binds to tubulin, preventing the action of microtubules during cell division 1jff anticancer DNA cisplatin binds to and causes crosslinking of DNA which ultimately triggers apoptosis (programmed cell death). 3lpv Cell signaling Beta-adrenergic receptor carazolol beta-blockers bind to and inhibits the adrenergic receptor used to reating heart disease 2rh1 Cell Signaling Prostaglandin H2 synthase aspirin inhibits the enzyme cyclooxygenase, which makes pain-signaling prostaglandins. 1pth Diseases of Affluence Pancreatic lipase alkyl phosphonate inhibits pancreatic lipase, reducing the amount of fat that is absorbed from food. 1lpb Diseases of Affluence HMG-CoA reductase Atorvastatin (Lipitor) inhibits HMG-CoA reductase, an enzyme involved in the synthesis of cholesterol. 1hwk B. Protein: Carbohydrate (CHO) interactions
Class Protein Carbohydrate Description PDB # Lectin – (CHO binding protein) Concavalin A dimannose Con A is from the Jack Bean and is used to study and purify carbohydrate binding proteinst 1I3H transporter Lactose permease Thiodigalactoside (a lactose analog) Escherichia coli lactose permease transports lactose , across cell membranes 1pv7 Human Lectin P-Selectin SLEX (a carbohydrate) SLeX, is a tetrasaccharidecarbohydrate that is found o the surface of cells. It is involved in cell-to-cell recognition and is one of the most important blood group antigens. P-selectin is a protein found on leukocytes, 1g1r C. Protein: Lipid Interactions
Class Protein Drug Description PDB # Binding insoluble fatty acids in cells Adipoctye lipid binding protein Arachadonic acid Intracellular protein involved in fatty acid biochemistry cyclooxygenase COX-2 (PDB code: 1CVU), 1adl Binding and transporting fatty acids in blood Human serum albumin stearic acid, Albumin is the most abundant protein in blood serum. It binds nonpolar molecules such as fatty acids 1e7i Cleave fatty acids from storage fats (triglycerides) Phospholipase A2 Octyl sulfate Releases fatty acid from triglycerides. Found in cells and is also secreted from cells 3fvi D. Protein: Small Molecule Interactions
Class Protein Drug Description PDB # Hormone Estrogen receptor Tamoxifen Tamoxifen, an estrogen mimic, is used to treat cancer by blocking the action of estrogen. 3ert Toxic molecules P-glycoprotein cyclic hexapeptide inhibitors, cyclic-tris- (R)-valineselenazole The protein is the most common molecular pump, removing toxic molecules (unfortunately also chemotherapeutic drugs) from cells. 3g60 Toxic molecules Cytochrome P450 Erythromycin (an antibiotic) CYP3A4 is the cytochrome p450 that plays the major role in drug detoxification and contributes to the metabolism of approximately 50% of marketed drugs. 2j0d Hormone Sex hormone binding globlulin testosterone testosterone is transported inside carrierproteins in the blood, including serum albumin and sex hormone binding globulin, 1d2s Hormone testosterone- binding domain tetrahydrogestrinone (THG) tetrahydrogestrinone (THG) is a synthetic designer anabolic steroid. 2amb Neurotrans Acetyl choline receptor Carbamoyl choline This brain neurotransmitter receptor is part of the nicotinic acid receptor system 1uv6 Neurotrans Human D3 Dopamine receptor eticlopride Structure of the human dopamine D3 receptor in complex with eticlopride, a dopamine antagonist 3pbl E. Protein: Metal Interactions
Class Protein ion Description PDB # transport Transferrin Iron Transferrin carries blood Fe, sequestering it and keeping it from oxidation to the more insoluble 3+ ferric form 1h76 storage Ferritin Iron Intracellular protein which form a shell that sequesters Fe ions 1fha Gene expression Gal 4 Zn Zn ions help stabilize a “Zn finger” structure in the protein, allowing it to bind to and regulate gene activity 1d66 Signaling cell to respond calmodulin calcium Calmodulin (calcium modulatory protein), on binding intracellular Ca ions, changes shape and causes cell responses 1exr F. Protein: Drugs of Abuse Interactions
Class Protein Drug Description PDB # Detection, Immunity Antibody THC A structural insight into the molecular recognition of a (- )-9-tetrahydrocannabinol and the development of a sensitive one-step, homogeneous immunocomplex based assay for its detection 3ls4 Drug activation, Toxin removal Human carboxylesterase 1 Cyclosarin (nerve agent) Human carboxylesterase 1 broad-spectrum bioscavenger that catalyzes the hydrolysis of acetyl groups from heroin (forming morphine) and cocaine, and the detoxification of organophosphate chemical weapons, such as sarin. 3k9b -
Choose a ligand to analyze
-
Select in turn hydrogen bond, hydrophobic, bridged H bond (mediated by a water molecule) and metal interaction. Take a cropped screenshot of each interaction.
-
For the final rendering, move the toggle on the Select Surfaces to opaque. Then change the distance in the best way to show the cavity in which the ligand binds. Color by hydrophobicity which gives two colors representing nonpolar and polar parts of cavity. Select solid surfaces.
-
Be prepared to explain in class the interactions shown in each rendering.
-
Cumulative Group Homework: Biochemistry’s Next Top Model
You have discovered a new protein isolated from a cell membrane. An initial check of protein databases indicates that your sequence is closely related to proteins that transport glucose across cell membranes.
Now you will need to gather other structural details based on its sequence. This lab will show you some of the things you can do with a computer, but also the limitations of structure prediction.
The primary sequence of the new protein is listed below in one letter codes. In this lab you will do some computer work and then try to build a physical model of the protein using wire.
There are four parts to the analysis.
Part 1: Hydropathy and Crossing a Membrane
Part 2: Secondary Structure Prediction
Part 3: Building a Model
Part 4: Tertiary Structure by Computer (Optional)
Group Member Names:
The Primary Sequence:
MEDDDGGLCLAVLVAMFFLGIMLLPSLANPPGRKGALLFAAIFCILAILMGVSAEG PKNRGALGIVLAAALCLVAIFGLRNQQSNSDGWPILLALTAQLAALCVLLLPFFPE SPRYQKKS
Part 1: Hydropathy and Crossing a Membrane
Most membrane bound proteins cross the cell membrane several times, so your first step is to construct a hydropathy plot of the protein. This will let you estimate how many times the protein crosses the cell membrane. The hydropathy scale below gives a positive score to hydrophobic side chains and negative scores to hydrophilic side chains.
-
Make a simple hydropathy plot:
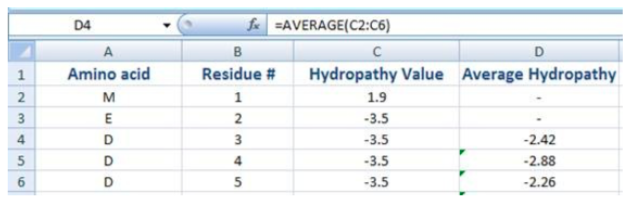
To make a simple hydropathy plot open an MS Excel file you have been sent named Next_Top_Model_Data. Column 1 has the amino acid sequence and column 2 has the residue #. Add hydropathy scores to column 3 and then make a ‘Column Graph’ in 2D format.
Hydropathy plots use an average value of the residues before and after the residue in question. In the example below the average hydropathy score for the aspartic acid in cell C4 is an average of the scores from cells C2 to C6. You can type ‘=average(C2:C6)’ to have Excel make an average in cell D4. Next, drag the bottom right corner of cell D4 all the way down the column. (For residues 1 and 2 will have no average value.) You will now have an average hydropathy for each residue that encompasses that residue plus the two before it and the two after it. This data will be used for graphing purposes.
Make a ‘Column Graph’ in 2D format of the Average Hydropathy (y-axis) and Residue #. Excel may plot the row numbers as well, so you may have to right click the graph and delete Series 1 or 2 under ‘Select Data’. Be sure that the residue # is on the x-axis. (In Excel 2007 you may need to go to the ‘Design’ panel and click ‘Switch Axes’.)
Answer these questions based on your plot:
-
From your hydropathy plot how many times does the protein appear to cross the cell membrane? (Consider what positive and negative values might indicate.)
-
About how many amino acids does it appear to take to cross a membrane?
-
If the N-terminus of the protein is inside the cell, where are residues 30-35 located?
-
Is the C-terminus intracellular or extracellular?
-
Considering what you know about the general position of side chains and the main chain atoms in a-helices and b-strands, do you believe that the portions that cross the membrane will most likely be a-helices or b-strands? (Note: This question will be answered later, so just make an educated guess/hypothesis here.)
Optional: The literature usually shows XY-scatter plots with a line connecting the dots for hydropathy. They are more common, but harder to read.
Amino Acid Hydropathy Scores
Amino Acid Name One Letter Code Hydropathy Score Isoleucine I 4.5 Valine V 4.2 Leucine L 3.8 Phenylalanine F 2.8 Cysteine C 2.5 Methionine M 1.9 Alanine A 1.8 Glycine G -0.4 Threonine T -0.7 Tryptophan W -0.9 Serine S -0.8 Tyrosine Y -1.3 Proline P -1.6 Histidine H -3.2 Glutamic acid E -3.5 Glutamine Q -3.5 Aspartic acid D -3.5 Asparagine N -3.5 Lysine K -3.9 Arginine R -4.5 Hydropathy Score Reference: Kyte, J. and Doolittle, R. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105-132.
-
Part 2: Secondary Structure Prediction
Predicting the 3D structure directly from a primary protein sequence is ‘the holy grail’ of biochemistry. It can currently be done only if the structure of a very similar protein is known. Secondary structural elements can be predicted with relative accuracy, however, since studies have shown that certain amino acids are more likely to be in a-helices or b-strands. In this portion you will take your sequence and predict some secondary structural elements. Go to the SCRATCH website which has programs that perform many functions related to proteins. Once there enter your e-mail address, the sequence listed in the first part of the lab, and then click the SSPRObox. In a few minutes you will get an e-mail with a code by each amino acid residue you submitted. (It might go to your Junk Mail.)
The SSPRO output has three possible codes. Paste the output below.
H = Helix
E = Extended beta structure
C = Turns, loops, and undefined structures
Answer these questions based on your secondary structure prediction:
-
How do the likely transmembrane segments of the sequence match up with helical segments from the SCRATCH program? (It may help to paste the results from SCRATCH below the primary sequence on page 1.)
-
Use the facts below about a-helices, and your data, to estimate the thickness of a cell membrane.
An \(\alpha\)-helix makes a turn every 3.6 residues. The pitch or advance per turn is 0.54 nm (nanometers).
-
Residues 30-33 (PPGR) seem to connect helix 1 and helix 2. Based on the number of residues between helices 1 and 2 what type of structure do residues 30-33 most likely represent?
-
Helix 1 ends at about residue 30 where there is a proline. Explain why it makes sense that proline would appear near the end of an a-helix. (Hint: Think about the H-bonding pattern.)
-
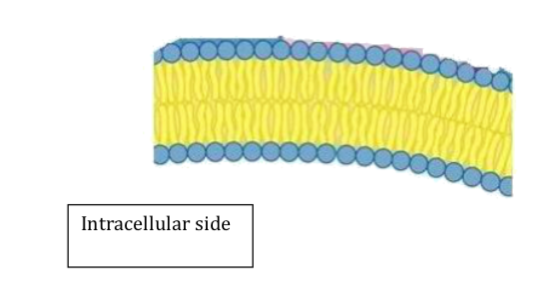
Shown below is a membrane bilayer (assume the area below it is inside the cell). Copy the picture and load it into the program Paint on your computer. (The program is in ‘Accessories’ on the list of programs.) In Paint choose the paint brush with a medium sized dot and draw the protein in with your mouse. Try to show about 5 turns per helix as they span the membrane. When done save your file and paste it back into this document. You can delete the original.
Part 3: Tertiary/3D StructurePart 3: Tertiary/3D Structure
As noted above 3D structure prediction is really challenging since residues far apart in the primary sequence may end up next to each other in the 3D structure. In this part of the lab you will attempt to build a physical model of your protein using a wire and various other supplies. The best looking model will get an awesome prize! Some assumptions for the model are:
-
Assume each amino residue is 1 inch long. Since the sequence has 120 amino acids you will need a 120 inch (10 foot) piece of wire. there are several types available. Note that some have somewhat sharp edges.
-
Color the N-terminus Blue and the C-terminus is red. (adding tape over maker will prevent smearing)
-
If you build an \(\alpha\)-helix one turn of the helix should be 3.6 inches of wire. The H-bonds that connect the turn in an \(\alpha\)-helix should be ~0.6 inches apart and go from 'loop to loop' in the same helx. try some short wires to add some H-bond examples.
-
For loops and extended structures not in the cell membrane just stretch out the wire with each residue being 1 inch in length.
-
The sequence also has 4 cysteine residues that appear to be in disulfide links. The approximate cysteine positions in helix are listed below. Use ashort red wire to make the most logical disulfide links.
Helix # 1 → Residue 4
Helix # 2 → Residue 10
Helix # 3 → Residue 10
Helix #4 → Residue 16
-
Take a look at how compacted your structure gets by measuring the distance between the N and C-termini. The wire is 10 feet if stretched out.
Part 4: Tertiary Structure Prediction (Optional)
As a final optional piece of the lab you can try a 3D shape predictor provided by the SCRATCH website. Go to the SCRATCH website and add your e-mail address and paste in the sequence from the beginning of the lab. This time click the 3Dpro box and you will get an e-mail with a computer prediction of the 3D structure. (This might take up to 30 minutes and the structure will be an attachment.) Once you get the structure you can move it with your left mouse button and your right mouse has options for color etc.)
Note: If the protein does not display correctly right click the structure and under ‘Display’ choose ‘Backbone’.
IV. Base Pairs -> DNA Helices
DNA is a polymer made of nucleic acid monomers.
Nucleic Acids:
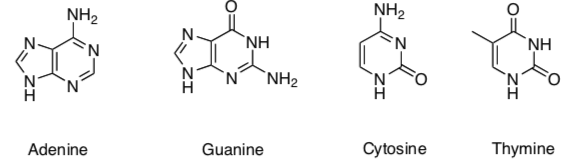
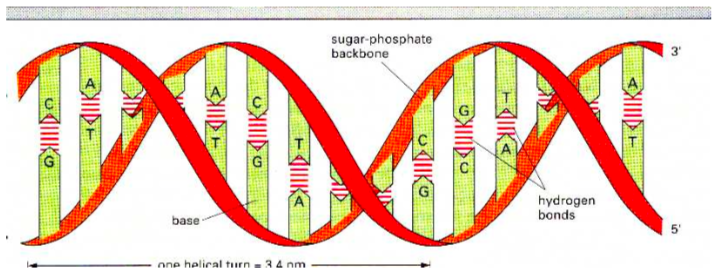
DNA is composed of a pair of single strand polymers that are held tightly together in the shape of a double helix.
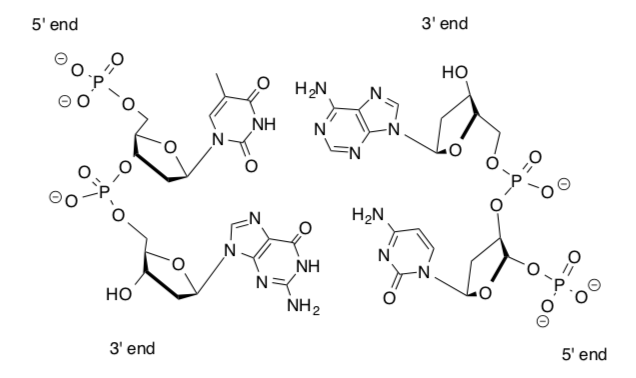
Think of the double helix as a twisted ladder. The sides consist of a repeating sugar-phosphate motif and the rungs between the strands are base pairs.
DNA is a stable molecule stabilized by several intermolecular forces.
-
Draw in the hydrogen bonding between these two strands of DNA.
-
Which bases “pair” with one another in DNA?
-
How many hydrogen bonds are formed between each of these pairs?
-
Define 3’ and 5’.
AG and CT bases pairs don’t typically occur. Let’s look at a few reasons.
Picture DNA as a ladder (below).
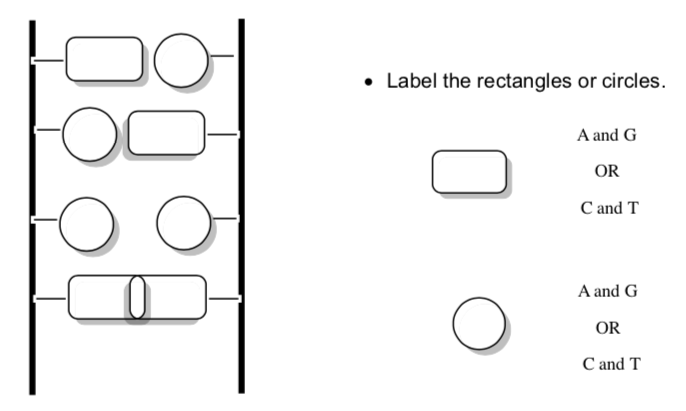
-
There are two problems AG and CT pairs using the ladder picture:
-
A and G are too [big or small] leading to _______________strain.
-
C and T are too [big or small] leading to weaker _____________. Think about Coulomb's law.
-
In terms of hydrogen bonding:
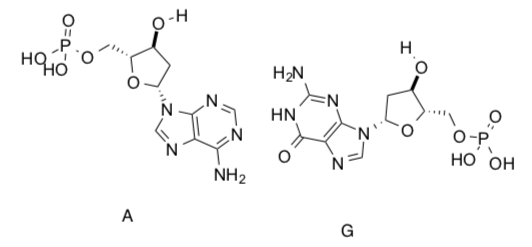
-
Compare the number of hydrogen bonds in the A:G base pair to the number of hydrogen bonds in the C:G base pair (previous page).
-
Which would be more stable?
DNA Base Pair Application Problem
-
A cyanophage is a virus that infects cyanobacteria.
In cyanophage S-2L, diaminopurine (DAP) is used instead of adenine. DAP basepairs perfectly with thymine forming 3 intramolecular hydrogen bonds.
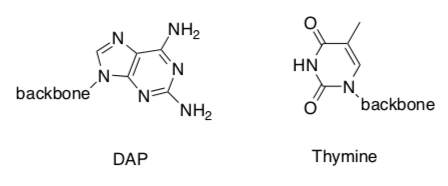
-
Identify the 3 hydrogen bonds.
A-T forms 2 hydrogen bonds. It has been suggested that the change to DAP from Adenine, helps the cyanophage virus to evade the bacterial host’s immune system.
-
Propose a reason for how DAP helps the virus survive.
-
-
Before Rosalind Franklin (and Watson and Crick) determined the structure of DNA, Chargaff noted that the total amount of A + G in DNA is equal to the amount of C + T, regardless of the type of DNA. Explain this observation.
-
Consider the single-stranded oligomer shown below:
-
Circle the phosphate functional groups.
-
Put a box around the sugar.
-
Draw the complementary strand showing the interactions between the base pairs.
-
Label the 3’ and 5’ ends.
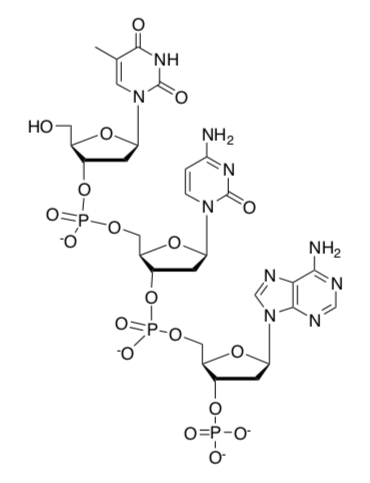
-
A single strand of DNA is an example of [ polymer or a supramolecular assembly ] ? Circle one.
-
A DNA double helix is an example of [ polymer or a supramolecular assembly ] ? Circle one.
-
Chiral DNA helix
Helices can be either right-handed or left-handed.
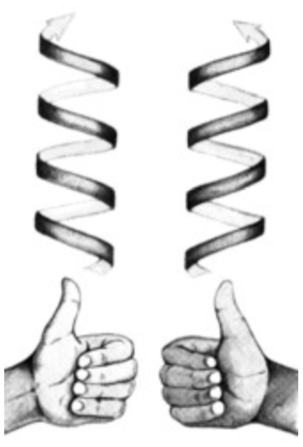
If you hold it pointing away from you and it twists clockwise moving away, it is right- handed, otherwise it is left-handed. These models are mirror images and can not be converted one into the other by rotation.
A and B forms of DNA are right-handed helices but Z-DNA is a left-handed helix.
-
Is the DNA in this picture a right-handed or left-handed helix?
Denaturation of DNA
The two strands of the double helix DNA are held together by relatively weak non- covalent interactions (hydrogen bonding and base stacking) and come apart quite easily known as denaturation. Denaturation caused by heating is called DNA melting. The temperature at which 50% of the DNA sample is denatured is calledmelting temperature (Tm).
-
Determine the melting temperature (Tm) for three DNA samples
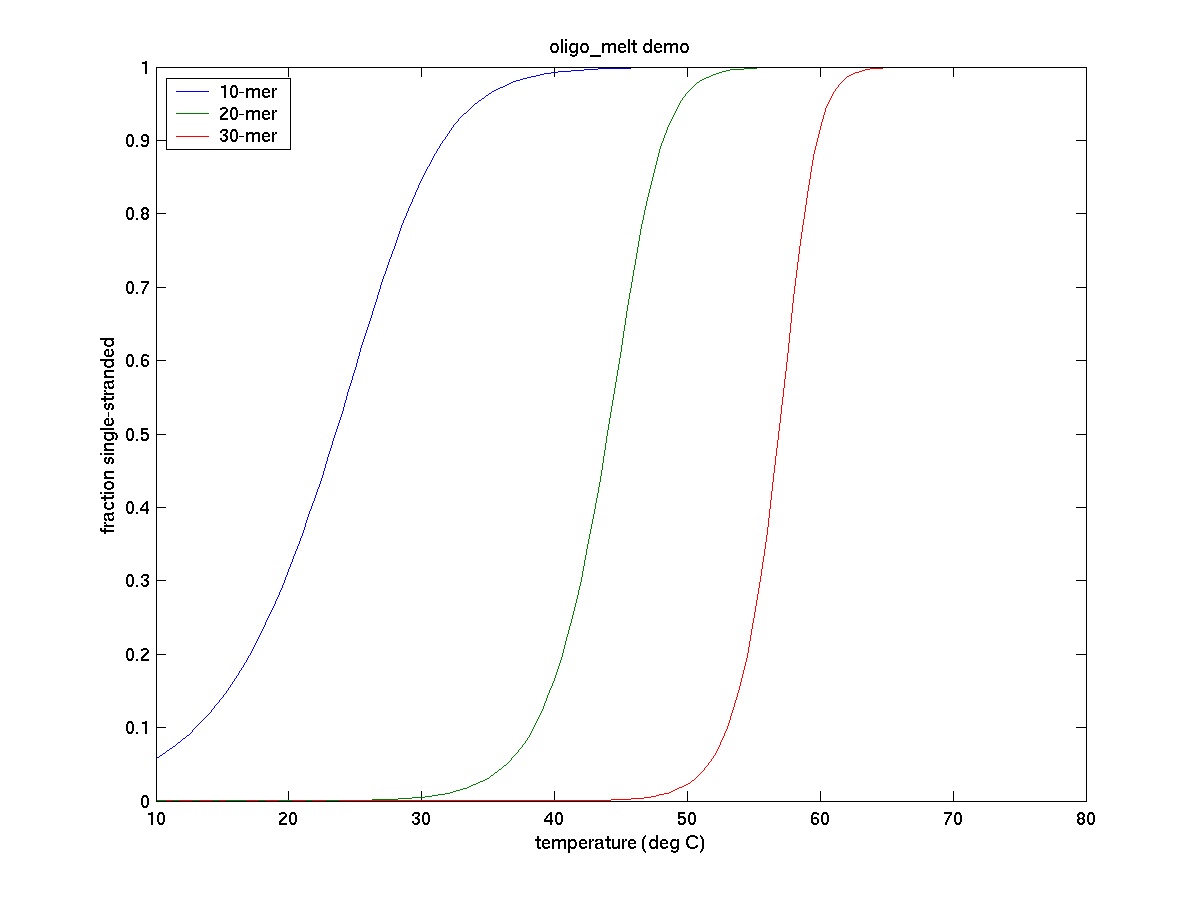
-
List all factors that would affect the Tm of a double helix DNA sample
Theoretical Tm
Recall that A:T base pairs have two hydrogen bonds and G:C base pairs have three. Given all other parameters are same, the melting point is determined by the G:C content of the DNA (the higher the G:C content the higher the temperature).
One very simple method to calculate theoretical Tm is to assign 20 C for each A:T pair and 4 0C for each G:C pair. The Tm is the sum of these values for all individual pairs in double stranded DNA.
-
Calculate the theoretical Tm for the following DNA
5’-ATATTACTGAATCAATAAT-3’
3’-TATAATGACTTAGTTATTA-5’
When the melting point of different DNA molecules are plotted against the molecule’s GC content a line is obtained as shown below:
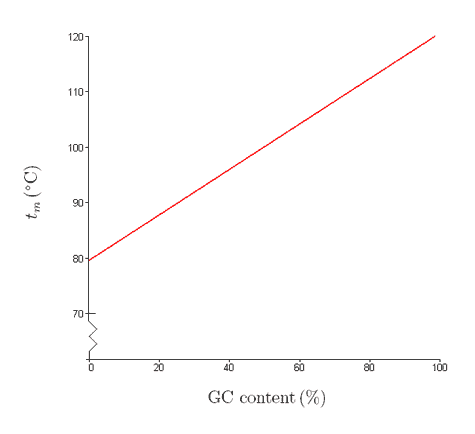
http://www.biology.arizona.edu/bioma...n/dnamelt.html
The linear equation used to calculate the melting point of a DNA molecule (0C, 100 mM Na+) is: Tm = 64.9 + 0.41* (%GC) – (500 /Number of base pair of DNA)
- Using the above equation, calculate the melting point of a DNA molecule that is 500 bp long and has 50% GC content.
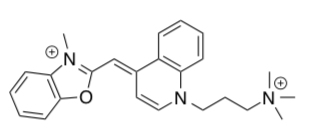
Major advances in single molecule research have been made possible with fluorescently labeled DNA. YO-PRO (structure shown below) is an oxazole dye that binds with double stranded DNA through “intercalation” (reversible inclusion between base pairs) mechanism. Recently researchers have reported that the melting temperature of a 30-base pair duplex DNA increased by ~20C for every intercalation site occupied by YO-PRO dye.
- Based on the structure of YO-PRO (shown below) propose an explanation for the increase of melting temperature (Tm).
DNA-Protein Complexes
DNA is huge and must be stored in the tiny nucleus of a cell. In eukaryotes, storing DNA is accomplished by binding it to small proteins called histones.
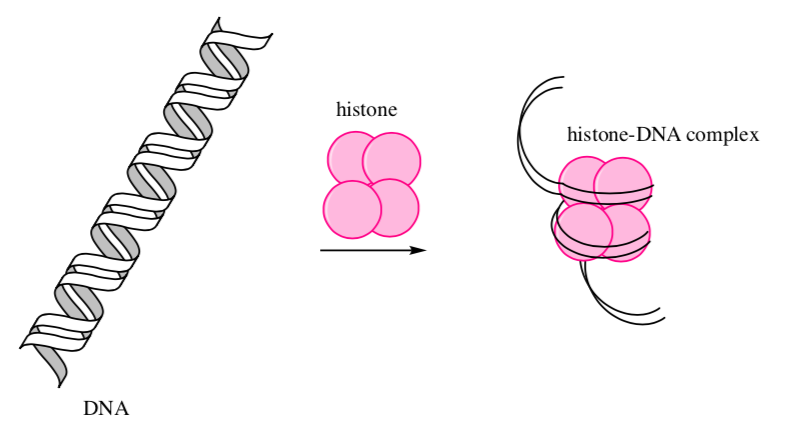
-
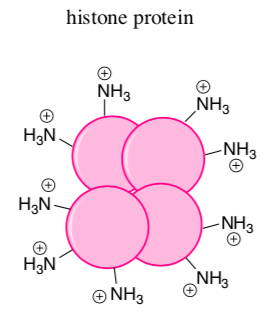
What is the overall charge on the DNA molecule? _______________
-
Since the histones form an ionic bond with DNA, predict what charge you would expect to see on the side chains of the histone proteins.
-
List some typical amino acids that would be involved.
-
Show an ionic bond between the histone and a DNA strand on the cartoon:
-
Would this binding depend on the sequence of base pairs in DNA?
DNA-Protein Complexes
Histones tightly bind to the DNA preventing transcription.
When it is time to transcribe DNA (to produce a needed protein), the histones need to unwind from the DNA.
Histones can be acetylated: acetyl (CH3CO) groups are attached to the amines.
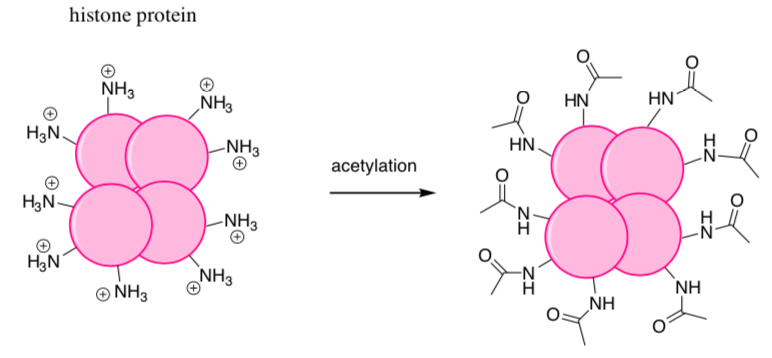
- Show why acetylation of the histone prevents it from interacting with DNA.
Additional Problems
DNA has a major and minor groove. To visualize these grooves, look at DNA at this site: http://employees.csbsju.edu/hjakubow...NAtutorial.htm
- Click on the the Spacefill Model - Major/Minor Grooves with O's (red) and N's (red with Hs not shown).
-
Some proteins bind through hydrogen bonding to DNA. Based on the model, where are there more hydrogen bond donors and acceptors?
Major Groove Minor Groove
Look at this model of a protein binding to DNA:
http://employees.csbsju.edu/hjakubow...REPRESSOR/Lamb da_Repressor.htm
- Draw the protein as a “cartoon” and show how it might interact through IMFs with DNA.
RNA
RNA, unlike DNA, uses ribose instead of deoxyribose and uracil instead of thymine as a base.
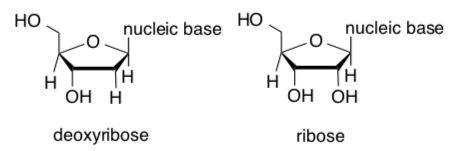
-
Clearly identify the difference(s) in these sugar structures.
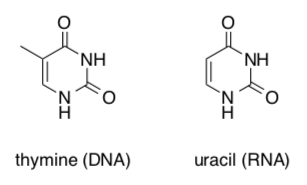
-
Clearly identify the difference(s) in these nucleic base structures.
-
The structure of thymine and uracil are shown above. Draw the structure of another base which could base pair with uracil in a short RNA
You have seen how drugs can bind through IMFs to the “active site” of a protein and inhibit the activity of the protein. A revolutionary new way to inhibit protein activity is to bind a molecule to RNAs that is decoded to form a given protein sequence. This prevents the protein from being made. If the “drug” is another RNA, this method is called RNA interference.
The top line below is part of a RNA sequence for a particular protein.
- Write underneath a sequence for a potential interfering RNA molecule.

RNA folding
Single stranded RNA can fold in on itself, like proteins, to form complicated “tertiary” structures. These contain short regions of “double-stranded” helices (“secondary structure”) stabilized by intramolecular H-bonds.
One common structure found in “folded” ssRNA is a stem/hairpin loop shown below.
-
Using the RNA base abbreviations: A, G, C and U, add RNA bases to the hairpin structure below which would allow a stable hairpin to form.

The loop in the stem/hairpin can serve a recognition site for a protein.
-
Draw a cartoon of a protein interacting with one loop of a folded ssRNA that has 3 different stretches of dsRNA.
RNA Application Problem
The longer the RNA, the harder it is to predict stem/hairpin structures since many are possible. Computer programs can do this for us.
Go to the mFold Web server at http://mfold.rna.albany.edu/?q=mfold
-
Select the RNA Folding Form link at the top left
-
Input a name for at test structure (use Test)
-
Enter the following single-stranded RNA sequence (listed in triplet pairs for ease of data input):
GAG ACA UCC AUU GAU ACA CGG GUU UUA UUU AUC AAU GGA UGU CUC
-
Select Format Sequence
-
Leaving all other choices at their default values, scroll down to the bottom and select FOLD RNA
-
In the output window select for Structure 1 an image file (jpg, png) and print it.
-
Make two different changes to the structure that would increase or decrease the stability of the hairpin. Rerun the analysis. Print both.
A chemical equation could be written for the folding of RNA (just as we did for the interconversion of substituted cyclohexane chair forms)
ssRNAunfloded ↔ RNAstem/loop
The ΔE for this reaction can be found at the bottom of the output file (dG = x). Use the following formula (as you did for chair interconversion) to find the Keq and the % folded;
ΔE (kcal/mol) = -1.4 log Keq
Summary of biomolecules
-
What are the four categories of biological molecules?
-
What are the polymers formed from each type of biological molecule?
-
Describe the structure of each category with pictures.
- Describe the role of IMFS in:
-
assembly of carbohydrates for storage in the liver
-
lipid aggregates
-
DNA double helix
-
Histone-DNA complex
-
secondary protein structures
-
substrate binding to an active site
-
multi-subunit protein (quaternary structure)
-
RNA hairpin
-

