7.6: Hydrogen Bonding
- Page ID
- 138869
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Methanol, CH4O (or CH3OH, a CH3 group attached to an OH), is another example of a molecule with a similar molecular weight to ethane. Like formaldehyde, methanol freezes somewhere around-90 oC, but it does not become a gas until it is heated to 65 oC. Methanol is certainly similar to formaldehyde in some ways. It contains oxygen and is very polar. The huge difference in their boiling points is due to the very strong hydrogen bonds in methanol.

Hydrogen bonding occurs when there is a significant amount of positive charge building up on a hydrogen atom. That happens because the hydrogen is attached to an atom that is much more electronegative than the hydrogen. As a result of that positive charge, a lone pair on another molecule strongly interacts with the hydrogen.
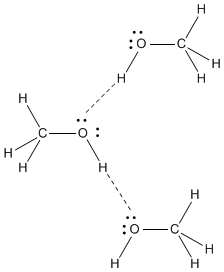
Hydrogen bonds occur only when a hydrogen is attached to one of the few most electronegative elements in the periodic table: fluorine, oxygen or nitrogen. It is these strong hydrogen bonds that are responsible for the relatively high boiling point of methanol; there is so much positive charge on the hydrogen of the OH group that it can essentially form a real bond with the lone pair on another methanol molecule.
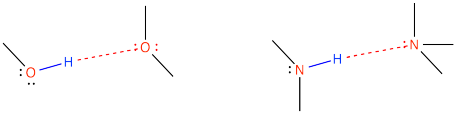
Water is an even better example of a hydrogen bonding compound; in fact, water molecules are so tightly bound to each other that this very small molecule freezes at 0 oC and does not boil until 100 oC (way, way hotter than an August day in Las Vegas). Water molecules are much less able to move around freely than are ethane molecules, even though ethane molecules weigh almost twice as much.
For practical purposes, hydrogen bonding usually involves compounds that contain N-H and O-H bonds. That's because oxygen and nitrogen are the second- and third-most electronegative elements in the periodic table, respectively. There are many, many examples of compounds like that. It also occurs in HF, of course, because fluorine is usually considered the most electronegative element, but there is really only one example of a compound with that bond, so you are much less likely to have to worry about that example. Slightly weaker hydrogen bonds have also been shown to occur involving other species, such as S-H bonds, but the most significant ones involve N-H and O-H bonds.
Problem SP6.1.
Which of the following molecules are capable of strong hydrogen bonding?
a) ethanal, CH3CHO b) ethylamine, CH3CH2NH2 c) acetic or ethanoic acid, CH3CO2H d) dimethyl peroxide, CH3OOCH3
Problem SP6.2.
Predict the relative order of melting points in the following amines:
hexylamine, CH3(CH2)4CH2NH2; triethylamine, (CH3CH2)3N; dipropylamine, (CH3CH2CH2)NH
Problem SP6.3.
Identify the strongest IMF in each case.

Biological Molecules and IMFs
Because compounds containing oxygen and nitrogen are so common in biology, hydrogen bonding is a fairly common feature in organic and biological chemistry.
Carbohydrates are one of the major classes of biomolecules. Carbohydrates are compounds that contain lots of OH groups, so they have lots of potential to form hydrogen bonds.
Proteins and peptides contain amide groups. Amides contain N-H bonds; hydrogen bonds involving amides are crucial influences in the structure of proteins.
Large biological molecules like proteins consists of 1000s of atoms, which interact through "intramolecular" ion-ion, London dispersion, dipole-dipole, and hydrogen bonding to create a unique 3D structure. Other molecules, both small and large, can bind to biological macromolecule using the same intermolecular interactions. An example of a protein, bovine low molecular weight protein tyrosyl phosphate, binding to inorganic phosphate, is shown below. Select the radio buttons below to alter the rendering of the molecular and obtain a better understanding of the structure/activity relationships of the protein:
|
spacefill |
wireframe |
cartoon |
| The displays above show three different ways to render the LMW-PTPase. You will recognize the spacefill and wireframe models, but note their complexity compared to the smaller molecules you have studied thus far. The cartoon molecules removes much of the detail by stripping away atoms and bonds, and replacing them with lines, ribbons (representing alpha helices) and block arrows (representing beta structure). | ||
|
cartoon with phosphate |
wireframe with phosphate |
|
| Inorganic phosphate, an inhibitor of this enzyme, binds to the enzyme through the intramolecular forces mentioned above. It is rendered in spacefill with the protein in cartoon and wireframe modes. | ||
DNA may be the most well-known example of a compound that displays hydrogen bonding. In DNA, complementary "base pairs" hydrogen bond to each other to held two strands of DNA together.
Weak, "Secondary" Hydrogen Bonds
Hydrogen bonding is really a special case of dipole forces. It occurs under very particular circumstances: in the presence of O-H or N-H bonds, which provide both a very positive hydrogen and a lone pair, the two features necessary for a hydrogen bonding interaction.
In general, we think of a hydrogen bond as the electrostatic interaction between an electron pair on one atom and a hydrogen atom that has built up a partial positive charge. There are some examples where that might be happenning, but that we would rather keep separate from the general idea of a hydrogen bond. For example, a lone pair on an oxygen atom in formaldehyde might be attracted to a hydrogen on another formaldehyde, which would be at the more positive end of the molecule. There is evidence from x-ray crystallography that such weak "hydrogen bonds" occur in the solid state in molecules containing aldehyde groups, similar to the one in formaldehyde.



