22: Nuclear Overhauser Effect (NOE)
- Page ID
- 332825
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Nuclear Overhauser Effect*
Nuclear Overhauser effect (nOe) is the transfer of nuclear spin polarization from one population of spin-active nuclei to another nuclei.
* All spectra in this unit are either from SDBS (Japan National Institute of Advanced Industrial Science and Technology) or simulated.
This transfer of energy results in a change in the intensity (positive or negative) of one NMR peak when another peak is irradiated with an RF field. Thus, we are seeing coupling from two nuclei that are close in space to each other.
An nOe is usually observed only on nuclei that are closer than 5 Å.
The most common application is for the determination of stereochemical and conformational relationships in relatively rigid molecules.
Examples:
A. There is restricted rotation around amide bonds. These specific nOe correlations could tell you which conformer was present.

B. These nOe correlations could tell you which E/Z isomer is present in an alkene. Draw the expected nOe correlations on the Z-isomer.
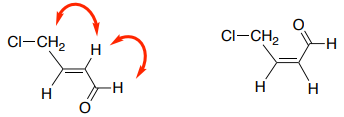
C. An nOe correlation could identify which stereoisomer is present. Draw the nOe correlations that you would use to determine which isomer is present.

nOe to Distinguish Isomers
The Park group completed this step in the synthesis of 1,6-Dideoxynojirimycin.
J.Org. Chem, 2008, 73, 2898.

- The authors noted a strong correlation between a quartet at 4.2 ppm and a vinylic proton at 6.4 ppm. Which isomer was formed?
More Practice:
- How could these pairs of isomers be distinguished using nOe correlations?


- What type of isomers are the pairs above? Circle one.
Enantiomers Diastereomers Conformers
- Could nOe correlations be used to distinguish enantiomers?
1D Experiment: nOe
To observe the nOe effect in a 1D experiment, a proton is irradiated and the effect on other proton signals is observed.

- How is this similar to the 1D spin decoupled experiment?
- How is this different from the 1D spin decoupled experiment?
nOe Difference Spectra
Because the nOe effect is often small, the original spectrum is subtracted from the irradiated spectrum to clearly show the differences.
- Use the NOE correlations to determine the correct structure. Draw arrows on the correct structure to indicate the nOe coupling between protons.


2D Experiments: NOESY and ROESY
In NOESY (Nuclear Overhauser Effect SpectroscopY) all correlations from nOe difference spectra are gathered in one spectrum.
The spectrum obtained is similar to COSY, with diagonal peaks and cross peaks, however the cross peaks connect resonances from nuclei that are spatially close rather than those that are through-bond coupled to each other.
In one of our first examples, we saw that nOe correlations could tell you distinguish between conformers of acetanilide.

- Use the NOESY data below to confirm which conformer of acetanilide was present.

NOESY in alpha-Helices of Proteins
In proteins, it is often important to determine the primary structure.

Scalar couplings (3JHH) can be used to determine the connectivity within an amino acid residue using COSY and TOCSY.
However, scalar couplings never occur between protons of different amino acids because of the amide bond.
NOEs can be used for that purpose. Since NOEs may be found between all protons close in space, this does not always work for sequencing.
In helical regions, sequential amide protons are close in space and NOEs will work.

- For the following helical section, determine the connectivity.
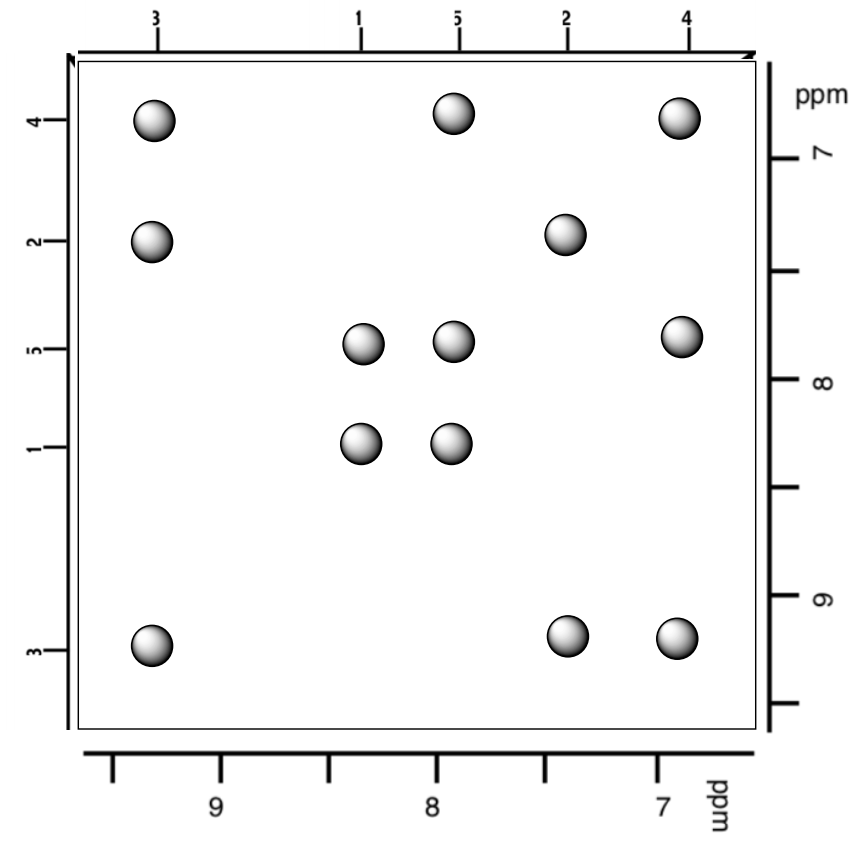
Practice Problem: Nepetalactone
Note: More NOE practice problems (and answers) are available at Graham, NMR2D [META: link missing]
Nepetalactone is the active ingredient in catnip. Use the following spectral data to determine the structure. Use the NOESY to determine the relative configuration.
Molecular Formula: C10H14O2 SU:____________
IR data 1746 & 1137cm-1 Functional Group: __________


| Number | 13C Shift | Interpretation |
|---|---|---|
| 1 | 170.13 | |
| 2 | 133.7 | |
| 3 | 115.4 | |
| 4 | 15.6 | |
| 5 | 40.7 | |
| 6 | 49.3 | |
| 7 | 30.9 | |
| 8 | 33.0 | |
| 9 | 39.7 | |
| 10 | 20.2 |
Practice Problem: Nepetalactone (cont.)

\(\boxed{\text{HMQC}}\)


| Number | 13C Shift | 1H Shift | Interpretation |
|---|---|---|---|
| 1 | 170.13 | ||
| 2 | 133.7 | ||
| 3 | 115.4 | ||
| 4 | 15.6 | ||
| 5 | 40.7 | ||
| 6 | 49.3 | ||
| 7 | 30.9 | ||
| 8 | 33.0 | ||
| 9 | 39.7 | ||
| 10 | 20.2 |
Practice Problem: Nepetalactone (cont.)
\(\boxed{\text{COSY}}\)


- Proposed Structure (Show Correlations):
Practice Problem: Nepetalactone (cont.)
\(\boxed{\text{NOESY}}\)

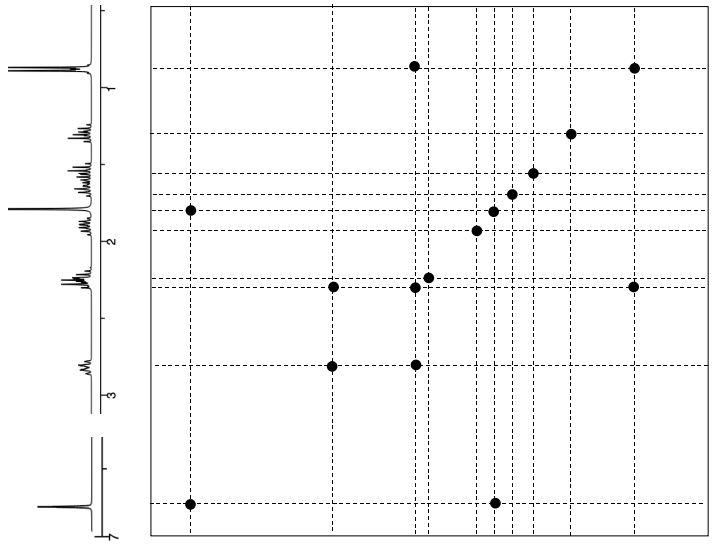
- Proposed Relative Stereochemistry (Show correlations with arrows)*
*Correct Answer: Google “nepetalactone”


