6: Mass Spectrometry
- Page ID
- 332806
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction to Mass Spectrometry
MS Experiment
Mass spectrometry (MS) is an instrument that measures the mass of charged particles.
In this instrument, the structure is hit with a high energy electron which causes the loss of an electron from the original structure.

Molecular Ion
The mass spectrometer sorts the ions produced based on their mass-to-charge ratio and a peak shows up for any positively charged ion.
- What would be the largest m/z peak for this ion?

- This first ionization is called the molecular ion. Does this match the molecular weight of the original compound?
Molecular Weights
Most organic compounds have even molecular weights.
However, If a sample has an odd number of nitrogens the molecular ion must be an odd.
- What is the MW for these compounds?

Isotopic Abundance in Mass Spectrometry*
Most elements exist as mixtures of different isotopes. Since a mass spectrometer separates and detects ions of slightly different masses, it easily distinguishes different isotopes of a given element.
Isotopes don’t impact the mass spectra of many organic compounds.
- The abundance of 12C is _________%.
- The abundance of 1H is _________%.
- The abundance of 16O is _________%.
However, compounds containing bromine and chlorine exhibit two molecular ions as they both have two very abundant isotopes.
Chlorine: 75.77% 35Cl and 24.23% 37Cl
Bromine: 50.50% 79Br and 49.50% 81Br
The mass spectrum of C6H6Br is shown below.
- Explain the two peaks at 156 and 158 m/z.
- Predict the MF (molecular formula) for this compound.

* Spectra in this unit are from SDBS (Japan National Institute of Advanced Industrial Science and Technology).
Practice
- The compound that gave this mass spectrum probably contains:
- nitrogen
- bromine
- chlorine
- sulfur

- The compound that gave this mass spectrum probably contains:
- nitrogen
- bromine
- chlorine
- sulfur
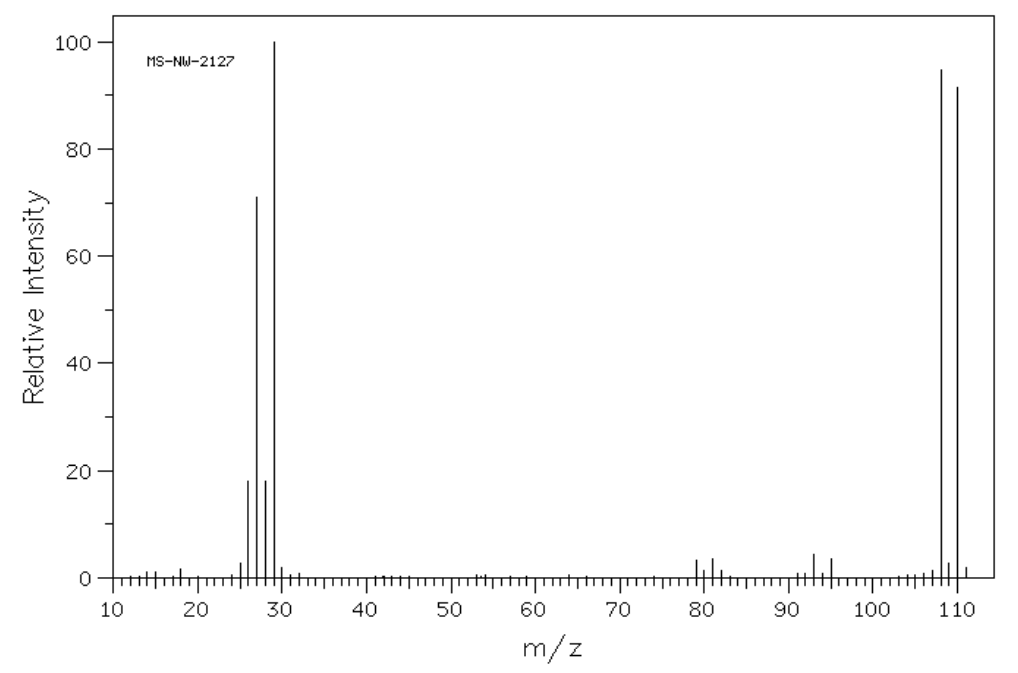
- How many molecular ions (compounds with different weights) would you expect to see in a molecule with 2 bromines? Explain.
Table of Molecular Ions Containing Br or Cl

- Using this table, propose a molecular formula for the compound that produced this spectrum.

Fragmentation in Mass Spectrometry
Initial Ionization
The radical cations (molecular ions) quickly decompose via radical reactions into fragments. Sometimes, the molecular ion is not even observed.
To start predicting fragments, you need to know the structure of the molecular ion.
- When a compound is ionized in the mass spectrometer, which electrons would be removed first? Explain your reasoning.

Review: Cation Stability
Usually a molecule will fragment to form the most stable cation. This cation will be most abundant and is called the base peak.
- Rank the following cations in order of increasing acidity and explain your ranking:

- Rank the following cations in order of increasing acidity and explain your ranking:

Fragmentation Mechanisms
Inductive Cleavage
- Show loss of an electron from the following molecule. Also add arrow(s) in the second step.

- Circle any ions that would be observed.
- What is the mass of the product(s) observed?
- Why does a bond break easily after this ionization?
Induction cleavage is also possible in the following pathways.
- Show the results of the first ionization and show the inductive cleavage that results, with arrows.

- In each case, give reasons why the bond broke easily.
Alpha Cleavage
- Show loss of an electron from the following molecule.
- Also add arrow(s) in the second step.

- Circle any ions that would be observed.
- What is the mass of the product(s) observed?
- Why does a bond break easily after this ionization?
Alpha cleavage is also possible in the following pathways.
- Show the results of the first ionization and show the alpha cleavage that results, with arrows.

- In each case, give reasons why the bond broke easily.
McLafferty Rearrangement
- Show loss of an electron from the following molecule.
- Also add arrow(s) in the second step.

- Why does the molecule rearrange after this ionization?
- What is the mass of the cationic fragment? Is it even or odd?
- What is the mass of the cationic fragments from alpha cleavages? Are they even or odd?
McLafferty rearrangement is also possible in the following pathway.
- Show the results of the first ionization and show the inductive cleavage that results, with arrows.

- What structural requirements are necessary for a McLafferty rearrangement?
Sigma Cleavage
- Show loss of an electron from the indicated sigma bond in the following molecule. Why does that bond then break easily?

- What is the molecular weight of the ion that results?
- Why is loss of an electron from a sigma bond less likely in the first place?
- Although impossible in the molecule shown above, what electrons are most likely to be lost in the initial ionization in mass spectrometry?
Summary of Mass Spectrometry
- The mass spectrometer can provide a molecular mass for any [ positively / negatively ] charged molecular ion or fragment.
- Mass spectrometry bombards a molecule to cause the loss of an electron and forming a molecular ion.
- The molecular ion is a [ radical / cation / anion / radical cation / radical anion ].
- This molecular ion is [ stable / unstable ].
- The molecular ion has the [ same MW / M+1 / M+2 / M-1 ] as the compound being measured.
- There are four major types of fragmentation. Show an example of each of these fragmentations.
- Inductive Cleavage:
- Sigma Cleavage:
- Alpha Cleavage:
- McLafferty Rearrangement:
Practice Fragmentation
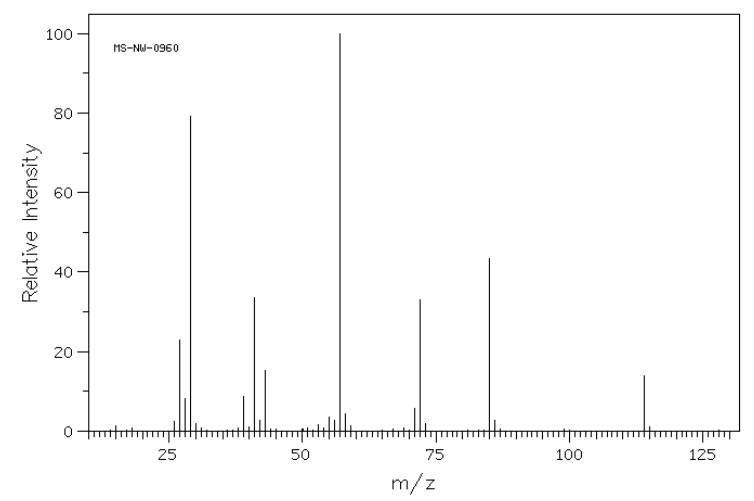
1. Analyze the following spectrum of 4-heptanone.
- Draw the structure of 4-heptanone.
- Show a mechanism for an alpha fragmentation and the McLafferty Rearrangement/Fragmentation for each compound.
- Use your predicted fragments to match the peaks within the MS.
2. Compounds X and Y are isomers with the formula C3H6O. Compound X has a peak at 1730 cm-1 in the IR and Y has a peak at 1715 cm-1. The spectra are shown below. Determine the structures of Compounds X and Y.
Compound X

Compound Y
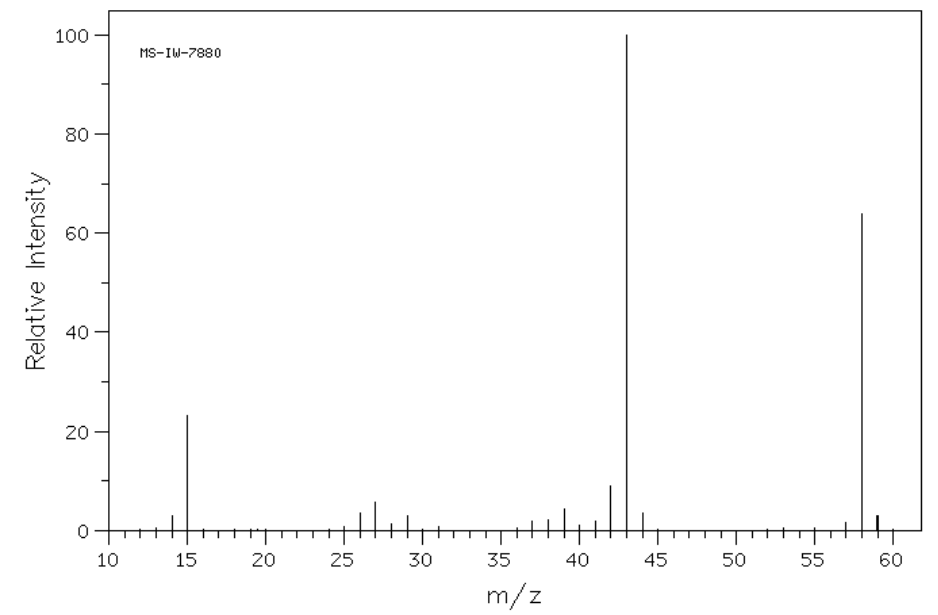
3. The mass spectra of three isomeric amines (sec-butylamine, isobutylamine and tert-butylamine) are shown below.
- Draw these three amines.
- Show all possible a-fragmentations.
- Match each spectrum with the correct structure.



4. Match the following mass spectra to the expected products. For each show how the assigned peaks were generated.

Compound A

- Show fragmentation that results in the peak at m/z = 43:
Compound B

- Show fragmentation that results in the peak at m/z = 44:
Compound C

- Show fragmentation that results in the peak at m/z = 57:
5. The following mass spectrum was obtained for compound D. Provide a mechanism for the formation of the base peak at m/z = 57.
Compound D 



