Urease 2
- Page ID
- 2516
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)
Introduction:
Urease is a metalloenzyme that catalyzes the reaction of urea into an ammonium ion and carbamate which then spontaneously breaks down to form ammonium and carbon dioxide. It was the first enzyme in history to be purified by James Sumner (1926). It is found in most soils as well as several plants including jack beans and soybeans. There are several different varieties of urease, found in plants, animals and microbes, with different chemical properties and compositions in each type.

Structure and Characteristics:
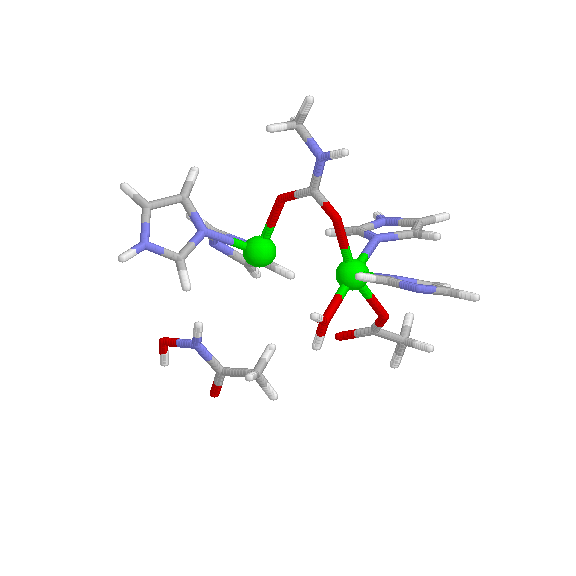
Urease is comprised of two half units which are held together by non-covalent bonds. Since the enzyme exists as a dimer, there are two active sites. Each active site is highly specific and will only bind to urea or hydroxyurea. The active sites of urease each contain a nickel atom, which is arranged in square pyramidal geometry with a coordination number of five. Each nickel atom has two imidazole nitrogen atoms, a carboxylate and a water molecule as ligands. They are connected by carbamate. The optimal pH of urease is 7.4. Its molecular weight is 480kDa. The urease enzyme has C2 symmetry, although there are asymmetrical states that exist only as a portion of the enzyme’s biological composition. IR spectroscopy can be used to identify urease, as it is IR active. Urease is inhibited by heavy metals such as lead complexes.
Reaction Rate and Environmental Effects on Reaction:
Urease activity is dependent upon the presence of urea and hydroxyurea substrates. As soon as the enzyme comes in contact with the substrate, it immediately catalyzes the decomposition of urea into ammonium and carbamate ions. Urease reactivity increases as temperature increases. Other factors such as organic matter and cation exchange capacity have an effect on urease activity.
History:
The ability of urease to hydrolyze urea into ammonium and carbon dioxide was discovered in 1909 by Takeuchi. Two years later, it was discovered that the enzyme was present in soybeans. In 1913, it was proposed that soybean urease could be applied to quantitatively determine presence of urea. Following the discovery of urease in soybeans, several experiments were performed to show that urease also exists in castor beans. These experiments showed that castor bean urease hydrolyzed less urea than soy bean urease, indicating that castor bean urease is less active than soybean urease. In 1926, Professor James B. Sumner was the first to discover that enzymes were proteins, by isolating and crystallizing urease from jack beans. At the time, it was believed that isolating an enzyme was impossible, and as a result, Sumner’s discovery was not accepted for years until another chemist named John H. Northrop was able to isolate an enzyme as well.
Practical Uses:
Urease is used to diagnose gastrointestinal diseases caused by the bacteria Helicobacter pylori , such as cancer or infections caused by the microorganism. The basic idea behind the test is that H. pylori bacteria are able to secrete urease and catalyze the reaction of urea into ammonium and bicarbonate. The test if performed by taking a biopsy of mucosa from the stomach and placing it in a medium containing urea and a type of indicator. The indicator is usually phenol red. If there are H. pylori bacteria present in the mucosa, the urease produced by the bacteria will hydrolyze the urea, raising the pH and causing a color change in the indicator from yellow to red. The color change is indicative of the presence of H. pylori due to its ability to produce urease. If there is no color change, no H. pylori is present in the sample. There are limitations to this test’s abilities to accurately diagnose gastrointestinal diseases, such as the fact that the bacteria move toward the center of the body in patients on certain drug therapies. In this case, biopsies would have to be taken from the fundus of the stomach as well as the antrum, instead of just the antrum in the normal test. Despite this limitation, the accuracy of the test is higher than other comparable urease testing methods, such as the urease breath test.
References:
1. Urease Fact Sheet. Mark S. Coyne Ph.D., Soil Microbiologist, Department of Agronomy, University of Kentucky. Web. <http://www.agrotaininternational.com...eFactSheet.pdf>.
2. Fishbein, William N., K. Nagarajan, and Warren Scurzi. "Urease Catalysis and Structure." Journal of Biological Chemistry 248.22 (1973): 7870-877.
3. Hubalek, Jaromir, Jan Hradecky, Vojtech Adam, Olga Krystofova, Dalibor Huska, Michael Masarik, Libuse Trnkova, Ales Horna, Katerina Klosova, Martin Adamek, Josef Zehnalek, and Rene Kizek. "Spectrometric and Voltammetric Analysis of Urease – Nickel Nanoelectrode as an Electrochemical Sensor." Sensors 4 (2007): 1238-255.
4. Austin, John W., Peter Doig, Murray Stewart, and Trevor J. Trust. "Structural Comparison of Urease and a GroEL Analog from Helicobacter Pyloni." Journal of Bacteriology 174.22 (1992): 7470-473.
5. Fishbein, William N., Thorne S. Winter, and J. D. Davidson. "Urease Catalysis." Journal of Biological Chemistry 240.6 (1965): 2402-406. Web.

