Urease 1
- Page ID
- 2515
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)
INTRODUCTION
In humans, urea is the main product of the metabolism and decomposition of nitrogen compounds. Urea has 30 ~ 40 kcal / mol of resonance energy1. Therefore, urea is a very stable. Moreover, in average, every person produces about 10 kg of urea per year, and the half-life of spontaneous hydrolysis of urea is about 3.6 years2. If nature does not have a valid decomposition process, urea will accumulate in the natural environment and cause serious problems. However, there is a hydrolase of urea -- urease exists in nature. Urease is able to decompose urea rapidly and produces ammonia and carbon dioxide (Figure 1).
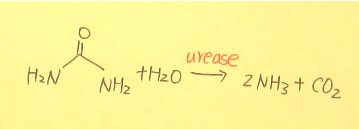 (Figure 1.)
(Figure 1.)
BACKGROUND
- Urease was the first enzyme to be crystallized. In 1926, a German scientist, Summer, successfully extracted some protein and crystallized a transparent square crystal from Canavalia ensiformis (Jack bean). The transparent square crystal was later approved urease. At that time, the X-ray crystal analysis technology is not that good yet , it cannot directly resolve the structure of protein crystals. Until 1995, Karplus then successfully used X-ray crystal technology to affirm the protein structure of urease3. Summer proved that protein is allowed to be crytallized. After 50 years of Summer’s observation of urease crytal, Blakeley and Zerner discovered the active site of urease has a metal, Ni, in the year of 1975.
- In particular, in many studies of hydrolase, urease is the only dinuclear hydrolase that containing nickel metal. Hence, by synthesized the organic and inorganic substances to get a mimicry compund which is similar to the urease active site is a very interesting study in the field of bioinorganic chemistry.
WHAT IS UREASE?
Urease and Agriculture
- Nitrogen, phosphorus and potassium are the three major nutrients to promote plant growth. Chlorophyl is necessary in the process of photosynthesis. The process of photosynthesis carries out product of glucose, then glucose furthering reacts with nitrogen to produce amino acids and protein.
- The lack nitrogen in plants will seriously affect the development of plant growing organs (roots, stems, leaves) especially the leaves part, so nitrogenous fertilizer is ofen referred as plant fertilizer. In addition, one of the most important source of nitrogenous fertilizer is urea. When farmers fertilize, whether the fertilizer can be absorbed and utilized by plants has become the key point of plant growing. Urease presents in many bacteria, fungi and higher plants. Its main function is to allow these organisms to gain nitrogen. However, the urease in the bacteria in soil will also cause the urea in fertilizer to be hydrolyzed, resulting in loss of fertilizer.
- From an agricultural experiment, scientists found that by adding urease inhibitor in fertilizer can increase the utilization of nitrogen in fertilizer for plants4. The common urease inhibitors are diphenols, quinones, hydroxamic acids, thiols and phosphoramides, etc.
Urease and Human
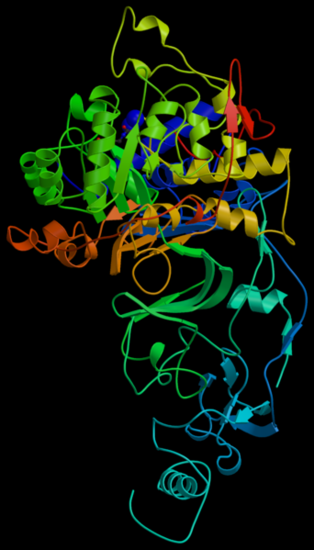
(Figure4.from Wikipedia) Molecular model of H. pylori urease enzyme
- Helicobacter pylori which is closely linked to human health also contains urease. Helicobacter pylori infection is one of the main causes that causes human pepticulcer. Helicobacter pylori infection may also cause Gastrointestinal disease like dyspepsia, chronic gastritis, gastric ulcer and duodenal ulcer, etc. It will even lead to more serious disease like gastric perforation and gastric cancer.
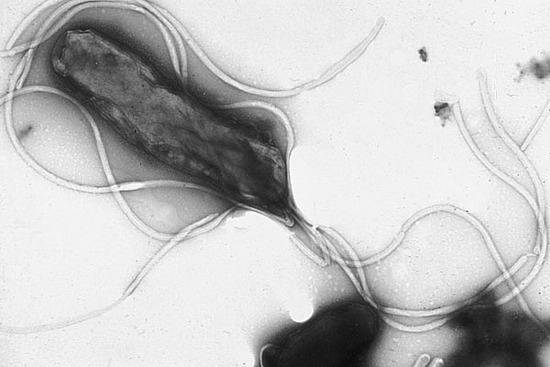
(Figure5.from Wikipedia) Helicobacter pylori
- Helicobacter pylori usually grows in surface of gastric mucosal layer. One of the reason why Helicobacter pylori is able to survive in environment of gastric acid is because the urease in Helicobacter pylori forms a layer on the surface of Helicobacter pylori in order to hydrolyze the urea into ammonia and use the ammonia to neutralize gastric acid. Therefore, because of the protecting surface, Helicobacter pylori is able to survive in the gastric acid environment.
ACTIVE SITE
- In 1989, the data from Wilcox’s SQUID showed that the urease active site is composed of two metal ions of high spin Ni (II) (S = 1), and between the two nickel-metal ions, there is weak antiferromagnetic exchange coupling. (J =-6.3cm-1)5
- In 1995, Karplus separated the urease from a bacteria called Klebsiella aerogenes; he successfully recrystallized the urease and gained the crystal structure of protein.From the study of X-ray structural analysis, urease is formed by three identical monomers known as ((αβγ)3). Each monomer contains an active site. In the X-ray structural analysis figure, people can see the two atoms is located at the place where the two nickel-metal-ion located on ative site. In average, molecular weight of each monomer are 83kDa (α = 60.3 kDa, β = 11.7 kDa, γ = 11.1 kDa).
- Active site of urease (Figure 2. and Figure 3.) is composed of two nickel-metal ions, the distance of between Ni ... Ni is 3.5 Å. Each nickel-metal ions has two Histidine residues and a water molecule. The two Ni metals are connected by a carbamylated lysine and hydroxide ion. Another Ni(2) has a monodentate ligand bond of Aspartate residues, making center of Ni (2) pseudooctahedral coordination geometry, while center of Ni (1) is pyramidal coordination geometry which has space in it.
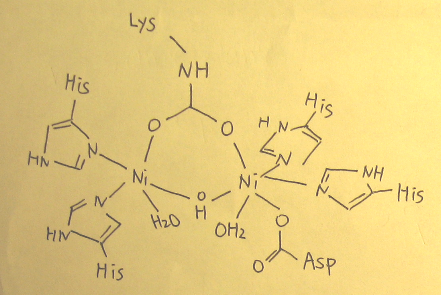
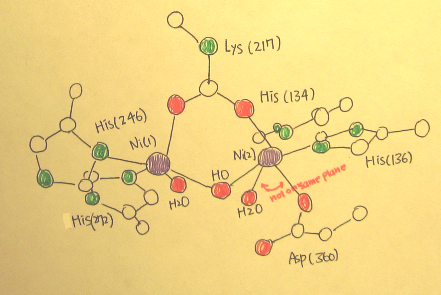
(Figure 2.) and (Figure 3.) - active site of Urease in crystallographic model
- The ligands to Ni (1) include6: 1. The second oxygen atom of the carbamylated Lys-217 2. Nitrogen atoms from His-246 and His-272 3. The partial coordination to the solvent molecule (H2O) is strongly coordinated to Ni (2).
- The ligands to Ni (2) include6: 1. Two nitrogen ligands which are derived from His-134 and His-136 2. Three oxygen atoms that are contributed by carbamylated Lys-217 and Asp-3603. Solvent molecule (H2O)
- Moreover, from the (Figure 3.) above we can see the active site of urease has no improper rotation axis, it must be dissymmetric. And there is no mirror plane because the two indicated bonds are not on the same plane. Therefore the point group for active site of urease is considered as C1.
- Number of symmetry elements: 1
- Number of irreducible representations:1
IR and RAMAN SPECTROSCOPY
Character Table
| C1 | E |
| A | 1 |
- There is no IR or Raman spectroscopy.
- IR Spectroscopy: Γvib must contain one of the x,y, z funtions to be observe. From the character table of C1, there is no x, y or z funtion to be observe. Hence there is no band in IR spectrum.
- Raman Spectroscopy: Γvib for urease (C1) does not contain one of x2, y2, z2, xy, xz, yx or combination such as x2+y2, x2-y2, 2z2-x2-y2. Therefore, there is no band in Raman spectroscopy.
REFERENCES
1. Wheland, G. W. Resonance in Organic Chemistry; Wiley:New York
2. Krebs, H. A.; Henseleit, K.; Hoppe-Seyler’s, Z. Physiol.Chem.
3. Zerner, B. Bioorg. Chem. 1991
4. Jabri, E.; Carr, M. B.; Hausinger, R. P.; Karplus, P. A.
5. Todd, M. J.; Hausinger, R. P. J. Biol. Chem. 1987
6. The Journal of Biological Chemistry, 271, August 2, 1996

