Ribulose 1,5-bisphosphate carboxylase (rubisco)
- Page ID
- 2511
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\) The limiting reaction within the Calvin Cycle is carried out by the enzyme Ribulose 1, 5-bisphosphate carboxylase. Considering the length of its name and the frequency of which it is talked about in modern science, the enzyme is commonly referred to as rubisco, (shown in figure two).1 This enzyme was discovered by Melvin Calvin at Berkeley in the 1940’s.2 Rubisco has since been named as the most abundant protein on earth. The enzymes relative abundance draws from its importance in nearly all plants and selected bacterium for its role in the fixation of carbon. Nearly all the biomass on earth has had contact with this very inefficient molecule.2
The limiting reaction within the Calvin Cycle is carried out by the enzyme Ribulose 1, 5-bisphosphate carboxylase. Considering the length of its name and the frequency of which it is talked about in modern science, the enzyme is commonly referred to as rubisco, (shown in figure two).1 This enzyme was discovered by Melvin Calvin at Berkeley in the 1940’s.2 Rubisco has since been named as the most abundant protein on earth. The enzymes relative abundance draws from its importance in nearly all plants and selected bacterium for its role in the fixation of carbon. Nearly all the biomass on earth has had contact with this very inefficient molecule.2
Rubisco in plants is a large protein with a molecular weight of 560,000 Dalton. The enzyme is located in the chloroplasts' stroma, consisting of at least 15% and up to 50% of the chloroplast protein.2 The high concentration of rubisco indicates a significant role for the plants metabolism. It is estimated that 4 x 1013g of the enzyme is made every second.3
Shape & Function
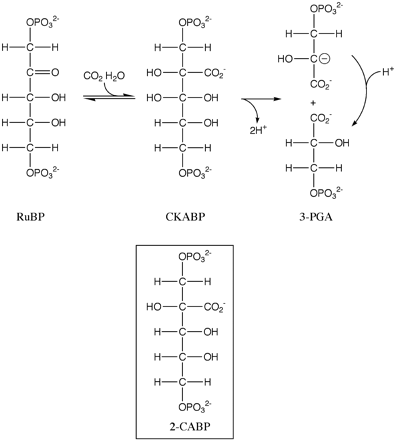 Rubisco’s shape directly relates to its function. In order for rubisco to function, it requires substrates supplied from the surrounding environment. These include carbon dioxide, Ribulose 1,5 bisphosphate (RuBP) and water, as depicted in figure 2.1 Once the substrates are gathered in specific location of the enzyme rubisco’s active site, the enzyme will perform a conformation change and essentially ‘slam’ the Carbon dioxide onto RuBP. The result is a highly unstable six-carbon reaction intermediate.1 Due to the molecules instability the carbon splits into two molecules of 3-phosphoglycerate (3GP). In the Calvin Cycle each of the 3GP molecules will be synthesized into Glyceraldehyde 3-phosphate (G3P). About one sixth of the G3P is used to make sugars, considered the output of the cycle.2
Rubisco’s shape directly relates to its function. In order for rubisco to function, it requires substrates supplied from the surrounding environment. These include carbon dioxide, Ribulose 1,5 bisphosphate (RuBP) and water, as depicted in figure 2.1 Once the substrates are gathered in specific location of the enzyme rubisco’s active site, the enzyme will perform a conformation change and essentially ‘slam’ the Carbon dioxide onto RuBP. The result is a highly unstable six-carbon reaction intermediate.1 Due to the molecules instability the carbon splits into two molecules of 3-phosphoglycerate (3GP). In the Calvin Cycle each of the 3GP molecules will be synthesized into Glyceraldehyde 3-phosphate (G3P). About one sixth of the G3P is used to make sugars, considered the output of the cycle.2
Rubisco is commonly considered the rate limiting step within the Calvin Cycle.4 It is largely inefficient considering the amount of energy it takes the average cell to make RuBP and its potential for photorespiration. Rubisco has a large active site that can accept both carbon-dioxide and O2 as a substrate. O2 has a higher affinity for the active site and therefore will bind at a higher rate than CO2.5 This inefficiency in the molecule has prompted different evolutionary traits from different species.6 Bacteria have developed carboxysome, small vesicles within the cell that have concentrated levels of rubisco.7 The carbon dioxide is shuttled from the outside environment in to the carboxysome to maximize the concentration exposure to rubisco. A more understood way of overcoming inefficiency problems comes from the plants of the world because of rubisco’s tendency to react with oxygen.
It is the molecules inefficiency and abundance that drives the bioscience world to rubisco’s possible improvement.8 Since all plants use the molecule, a protein that performs the carbon fixing function of rubisco and its active site is non-competitive would greatly reduce the amount of energy and nitrogen that a plant needs to build biomass.8 Such a protein has the potential to change the world from farming to the environment and even the carbon content in our atmosphere.9
Stereochemistry and Spectroscopy
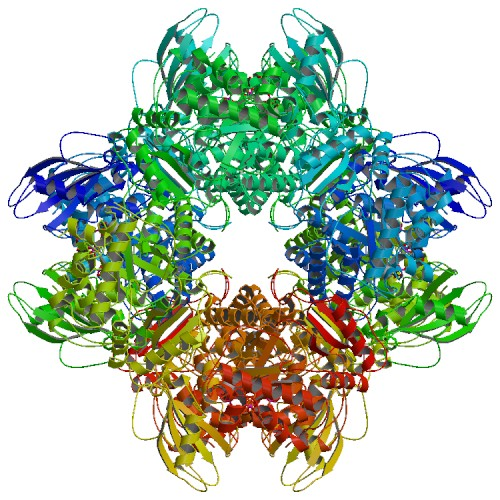 A first glance analysis of the entire enzymes crystal structure, as shown in figure 3, it looks as if it has C4 point group symmetry. The primary axis would be running straight down the center of the molecule. Further inspection and tying information form Figure 1 reveals that rubisco is large enzyme containing four protein strands that vary in collapsed shape. This fact is indicated in the color pattern of figure 3. Therefore it can be concluded by simple inspection that the overall structure is C1 point group. Because this molecule is large and contains many sets of atoms it is increasingly difficult to determine if the entire rubisco structure will be either infrared or Ramman active.
A first glance analysis of the entire enzymes crystal structure, as shown in figure 3, it looks as if it has C4 point group symmetry. The primary axis would be running straight down the center of the molecule. Further inspection and tying information form Figure 1 reveals that rubisco is large enzyme containing four protein strands that vary in collapsed shape. This fact is indicated in the color pattern of figure 3. Therefore it can be concluded by simple inspection that the overall structure is C1 point group. Because this molecule is large and contains many sets of atoms it is increasingly difficult to determine if the entire rubisco structure will be either infrared or Ramman active.
In the analysis of the active site within rubisco it has been studied that there is the involvement of Mg2+. This alkaline earth metal stabilizes formation of an enediolate carbanion equivalent that is subsequently carboxylated by CO2, as shown in figure 4.3 This conformation change, as shown in studies, activates a broadband of visible and infrared emissions. Once in stable formation Mg2+, although not well depicted in figure 4, will form covalent bonds with four Oxygen atoms. This formation can be described as square planar when considering only the immediate ligands around Mg2+. The point group, off of a character table, can be described as D4h. 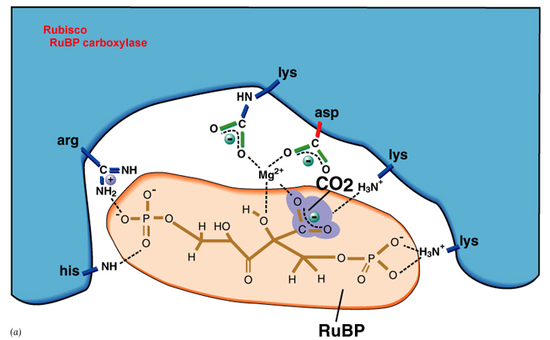
Continuing off of the inferred orientation, infrared spectrum using molecular orbital theory will show up at two bands and Raman will show three. Although no studies have been confirmed for Raman spectroscopy, there have been studies that show near-infrared emissions do show when the molecule has changed formation around the magnesium and its substrates.10 This conformation changes the overall structure of the rubisco enzyme only slightly. The change in the spectra can be rationalized to be form the Mg++ bonded to the active site and the substrates. To further this rationalization is has been showed that the shift in spectra scatters as RuBP concentrations become low in a limited culture environment.11
References
- Boyer, R., 2006 Synthesis of Carbohydrates by the Calvin Cycle, pp. 144-147 in Concepts in BioChemistry: Third Edition edited by K. MOLLOY. John Wiley & Sons, Inc., Hoboken.
- Vapaavuori, E. M. Correlation of Activity and Amount of Ribulose 1,5-<I>Bis</I>Phosphate Carboxylase with Chloroplast Stroma Crystals in Water-Stressed Willow Leaves. Journal of Experimental Botany 1986. 37, 174, 189-198.
- Lundqvist, T.; Schneider, G. Crystal Structure of Activated Ribulose-1,5-Bisphosphate Carboxylase Complexed with Its Substrate, Ribulose-1,5-Bisphosphate. Journal of Biological Chemistry 1991, 266, 12604-12611.
- Badger, M. R.; Price, G. D. Co2 Concentrating Mechanisms in Cyanobacteria: Molecular Components, Their Diversity and Evolution. J. Exp. Bot. 2003, 54, 609-622.
- Tabita, F. R.; Hanson, T. E.; Li, H.; Satagopan, S.; Singh, J.; Chan, S. Function, Structure, and Evolution of the Rubisco-Like Proteins and Their Rubisco Homologs. Microbiol. Mol. Biol. Rev. 2007, 71, 576-599.
- Badger, M. R.; Bek, E. J. Multiple Rubisco Forms in Proteobacteria: Their Functional Significance in Relation to Co2 Acquisition by the Cbb Cycle. J. Exp. Bot. 2008, 59, 1525-1541.
- Taylor, T. C.; Backlund, A.; Bjorhall, K.; Spreitzer, R. J.; Andersson, I. First Crystal Structure of Rubisco from a Green Alga,Chlamydomonas Reinhardtii. Journal of Biological Chemistry 2001, 276, 48159-48164.
- Spreitzer, R. J.; Salvucci, M. E. Rubisco: Structure, Regulatory Interactions, and Possibilities for a Better Enzyme. Annual Review of Plant Biology 2002, 53, 449-475.
- Parry, M. A. J.; Andralojc, P. J.; Mitchell, R. A. C.; Madgwick, P. J.; Keys, A. J. Manipulation of Rubisco: The Amount, Activity, Function and Regulation. J. Exp. Bot. 2003, 54, 1321-1333.
- Sadava, H., Orians, Purves, Hills. Radioisotope Labeling Experiments Revealed the Steps of the Calvin Cycle. In Life: The Science of Biology, The Courier Companies, Inc.: Gordonsville, 2008; Vol. Eight Edition, pp 170-172.
- Kannappan, B.; Gready, J. E. Redefinition of Rubisco Carboxylase Reaction Reveals Origin of Water for Hydration and New Roles for Active-Site Residues. Journal of the American Chemical Society 2008, 130, 15063-15080.

